Figure 1: GemCode single-cell technology enables 3′ profiling of RNAs from thousands of single cells simultaneously.
From: Massively parallel digital transcriptional profiling of single cells
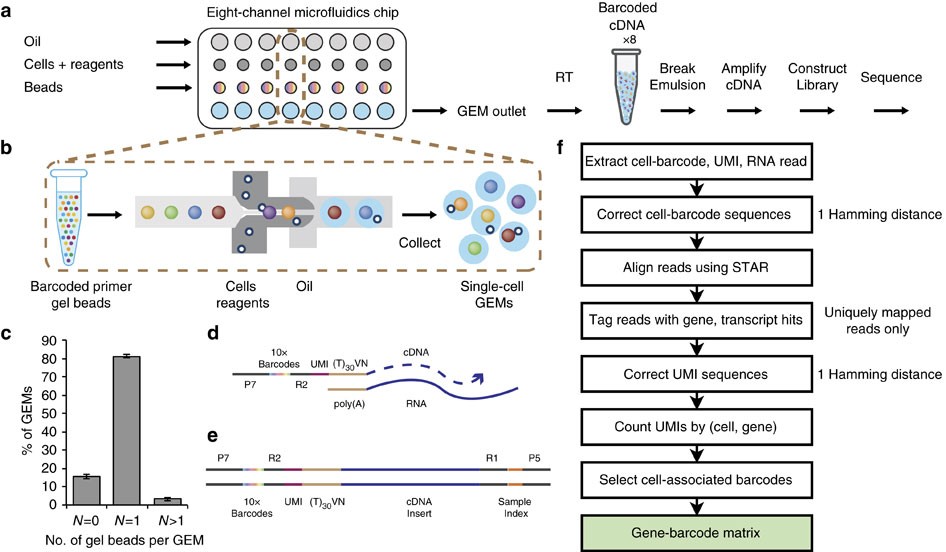
(a) scRNA-seq workflow on GemCode technology platform. Cells were combined with reagents in one channel of a microfluidic chip, and gel beads from another channel to form GEMs. RT takes place inside each GEM, after which cDNAs are pooled for amplification and library construction in bulk. (b) Gel beads loaded with primers and barcoded oligonucleotides are first mixed with cells and reagents, and subsequently mixed with oil-surfactant solution at a microfluidic junction. Single-cell GEMs are collected in the GEM outlet. (c) Percentage of GEMs containing 0 gel bead (N=0), 1 gel bead (N=1) and >1 gel bead (N>1). Data include five independent runs from multiple chip and gel bead lots over >70k GEMs for each run, n=5, mean±s.e.m. (d) Gel beads contain barcoded oligonucleotides consisting of Illumina adapters, 10x barcodes, UMIs and oligo dTs, which prime RT of polyadenylated RNAs. (e) Finished library molecules consist of Illumina adapters and sample indices, allowing pooling and sequencing of multiple libraries on a next-generation short read sequencer. (f) CellRanger pipeline workflow. Gene-barcode matrix (highlighted in green) is an output of the pipeline.
