Figure 1: Secondary oligos are specific and efficient.
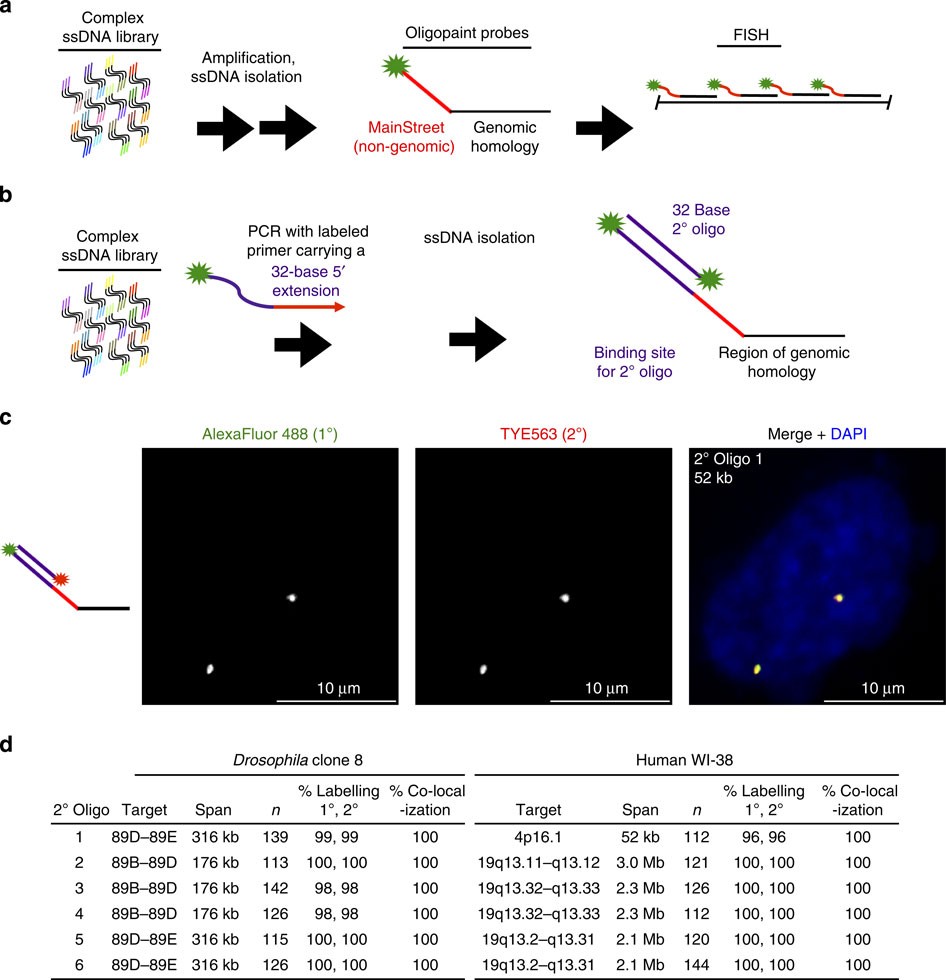
(a) One synthesis strategy for Oligopaints, in which complex ssDNA libraries consisting of a stretch of genomic sequence (black lines) on the order of tens of bases flanked by non-genomic regions (coloured lines) containing primer sequences are amplified, labelled and then processed in any of a variety of ways to produce ssDNA probes that carry non-genomic sequences at one (shown) or both (not shown) ends (adapted from ref. 7; also see Supplementary Figs 2 and 3 for more details on MainStreet incorporation and placement strategies). The primer sequence can constitute the entirety, or just a portion, of the non-genomic region, called MainStreet, which will remain single-stranded when Oligopaint probes are hybridized to their target. (b) A binding site for a secondary (2°) oligo probe can be introduced to MainStreet by PCR amplification with a primer that carries the binding site. Here, the secondary oligo carries a single, 5′ fluorophore that matches the fluorophore present on the Oligopaint (primary) probe, but in practice the number, identity and placement of fluorophores on the secondary oligo can vary. Also see Supplementary Figs 2 and 3. (c) Grayscale and multicolour images from a two-colour co-localization experiment in diploid human WI-38 cells. DNA is stained with 4′,6-diamidino-2-phenylindole (DAPI; blue). Images are maximum Z projections from a laser scanning confocal microscope. (d) Two-colour co-localization experiments in diploid Drosophila clone 8 cells and WI-38 cells. The genomic target, span of the target, number of nuclei examined (n), per cent of nuclei (% Labelling) that had at least one signal from the primary (1°, Oligopaint) probe and at least one signal from the secondary oligo and per cent primary signals that have an overlapping secondary signal (% Co-localization) are given for each experiment.
