Abstract
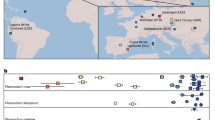
We sequenced and annotated the genomes of four P. vivax strains collected from disparate geographic locations, tripling the number of genome sequences available for this understudied parasite and providing the first genome-wide perspective of global variability in this species. We observe approximately twice as much SNP diversity among these isolates as we do among a comparable collection of isolates of P. falciparum, a malaria-causing parasite that results in higher mortality. This indicates a distinct history of global colonization and/or a more stable demographic history for P. vivax relative to P. falciparum, which is thought to have undergone a recent population bottleneck. The SNP diversity, as well as additional microsatellite and gene family variability, suggests a capacity for greater functional variation in the global population of P. vivax. These findings warrant a deeper survey of variation in P. vivax to equip disease interventions targeting the distinctive biology of this neglected but major pathogen.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
NCBI Reference Sequence
Sequence Read Archive
References
Guerra, C.A. et al. The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Negl. Trop. Dis. 4, e774 (2010).
Dobson, M.J. Malaria in England: a geographical and historical perspective. Parassitologia 36, 35–60 (1994).
Price, R.N., Douglas, N.M. & Anstey, N.M. New developments in Plasmodium vivax malaria: severe disease and the rise of chloroquine resistance. Curr. Opin. Infect. Dis. 22, 430–435 (2009).
Carlton, J.M., Sina, B.J. & Adams, J.H. Why is Plasmodium vivax a neglected tropical disease? PLoS Negl. Trop. Dis. 5, e1160 (2011).
Winzeler, E.A. Malaria research in the post-genomic era. Nature 455, 751–756 (2008).
Carlton, J.M. et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455, 757–763 (2008).
Dharia, N.V. et al. Whole-genome sequencing and microarray analysis of ex vivo Plasmodium vivax reveal selective pressure on putative drug resistance genes. Proc. Natl. Acad. Sci. USA 107, 20045–20050 (2010).
Gnerre, S. et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA 108, 1513–1518 (2011).
Feng, X. et al. Single-nucleotide polymorphisms and genome diversity in Plasmodium vivax. Proc. Natl. Acad. Sci. USA 100, 8502–8507 (2003).
Mu, J. et al. Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol. Biol. Evol. 22, 1686–1693 (2005).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Gemayel, R., Vinces, M.D., Legendre, M. & Verstrepen, K.J. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 44, 445–477 (2010).
Joy, D.A., Mu, J., Jiang, H. & Su, X. Genetic diversity and population history of Plasmodium falciparum and Plasmodium vivax. Parassitologia 48, 561–566 (2006).
Kumar, S. & Subramanian, S. Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. USA 99, 803–808 (2002).
Paget-McNicol, S. & Saul, A. Mutation rates in the dihydrofolate reductase gene of Plasmodium falciparum. Parasitology 122, 497–505 (2001).
Cornejo, O.E. & Escalante, A.A. The origin and age of Plasmodium vivax. Trends Parasitol. 22, 558–563 (2006).
Conway, D.J. et al. Origin of Plasmodium falciparum malaria is traced by mitochondrial DNA. Mol. Biochem. Parasitol. 111, 163–171 (2000).
Carter, R. Speculations on the origins of Plasmodium vivax malaria. Trends Parasitol. 19, 214–219 (2003).
Thera, M.A. et al. A field trial to assess a blood-stage malaria vaccine. N. Engl. J. Med. 365, 1004–1013 (2011).
Fernandez-Becerra, C. et al. Plasmodium vivax and the importance of the subtelomeric multigene vir superfamily. Trends Parasitol. 25, 44–51 (2009).
Bozdech, Z. et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc. Natl. Acad. Sci. USA 105, 16290–16295 (2008).
Garnham, P.C. et al. A strain of Plasmodium vivax characterized by prolonged incubation: morphological and biological characteristics. Bull. World Health Organ. 52, 21–32 (1975).
Nayar, J.K. et al. Studies on a primaquine-tolerant strain of Plasmodium vivax from Brazil in Aotus and Saimiri monkeys. J. Parasitol. 83, 739–745 (1997).
Bhasin, V.K. & Trager, W. Gametocyte-forming and non-gametocyte–forming clones of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 33, 534–537 (1984).
Sullivan, J.S. et al. Adaptation of a strain of Plasmodium vivax from India to New World monkeys, chimpanzees, and anopheline mosquitoes. J. Parasitol. 87, 1398–1403 (2001).
Collins, W.E. et al. Studies on the North Korean strain of Plasmodium vivax in Aotus monkeys and different anophelines. J. Parasitol. 71, 20–27 (1985).
Wellems, T.E. et al. Chromosome size variation occurs in cloned Plasmodium falciparum on in vitro cultivation. Rev. Bras. Genet. 11, 813–825 (1988).
Collins, W.E. et al. Adaptation of a strain of Plasmodium vivax from Mauritania to New World monkeys and anopheline mosquitoes. J. Parasitol. 84, 619–621 (1998).
Gardner, M.J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 (2002).
Melnikov, A. et al. Hybrid selection for sequencing pathogen genomes from clinical samples. Genome Biol. 12, R73 (2011).
Birney, E., Clamp, M. & Durbin, R. GeneWise and Genomewise. Genome Res. 14, 988–995 (2004).
Lagesen, K. et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108 (2007).
Lowe, T.M. & Eddy, S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Griffiths-Jones, S. Annotating non-coding RNAs with Rfam. Curr. Protoc. Bioinformatics Chapter 12, Unit 12.5 (2005).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Ter-Hovhannisyan, V., Lomsadze, A., Chernoff, Y.O. & Borodovsky, M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18, 1979–1990 (2008).
Blanco, E. & Abril, J.F. Computational gene annotation in new genome assemblies using GeneID. Methods Mol. Biol. 537, 243–261 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Kolpakov, R., Bana, G. & Kucherov, G. mreps: efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res. 31, 3672–3678 (2003).
Russell, B., Suwanarusk, R. & Lek-Uthai, U. Plasmodium vivax genetic diversity: microsatellite length matters. Trends Parasitol. 22, 399–401 (2006).
Ina, Y. New methods for estimating the numbers of synonymous and nonsynonymous substitutions. J. Mol. Evol. 40, 190–226 (1995).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Acknowledgements
This work has been funded, in whole or in part, by federal funds from the US National Institute of Allergy and Infectious Diseases (NIAID), the US National Institutes of Health (NIH) and the US Department of Health and Human Services, under contracts HHSN266200400001C and HHSN2722009000018C. We gratefully acknowledge the Indian Council of Medical Research for financial support of the Malaria Parasite Bank at the National Institute of Malaria Research, New Delhi, and the NIMR Director for providing all facilities. We thank the NIAID/National Human Genome Research Institute (NHGRI) Eukaryotic Pathogens and Disease Vectors Working Group and the Broad Institute Genome Sequencing Platform for significant contributions to this project. K.G. and L.Y. are supported by a Global Health Program grant from the Bill and Melinda Gates Foundation. A.A.E. is supported by a grant from the NIH (RO1GM084320), and J.M.C. and P.L.S. are supported by grant U19AI089676, an NIAID International Center of Excellence for Malaria Research. The content is soley the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author information
Authors and Affiliations
Contributions
J.W.B. provided P. vivax strains, and A.R.A. and A.P.D. provided Indian P. falciparum material. S.M.S., S.S. and S.Y. performed genome assembly. S.G., J.M.G. and Q.Z. performed genome annotation. K.G., R.H.Y.J., L.Y. and D.E.N. performed analyses. S.B.C. performed project management. P.L.S. undertook experimental validation. D.E.N., A.A.E., B.W.B. and J.M.C. directed the analyses. D.E.N., A.A.E., J.W.B. and J.M.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary Information for
41588_2012_BFng2373_MOESM19_ESM.xls
Comparison of nonsynomymous:synonymous diversity ratio (πN/πS) in P. vivax and P. falciparum orthologs (.xlsx file) (XLS 136 kb)
41588_2012_BFng2373_MOESM20_ESM.xls
Gene categories differentially enriched (Z > 2) for functional diversity (πN/πS) in P. vivax or P. falciparum (.xlsx file) (XLS 34 kb)
Rights and permissions
About this article
Cite this article
Neafsey, D., Galinsky, K., Jiang, R. et al. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet 44, 1046–1050 (2012). https://doi.org/10.1038/ng.2373
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ng.2373
This article is cited by
-
Comparative genomic analysis of Babesia duncani responsible for human babesiosis
BMC Biology (2022)
-
A genetically modified Plasmodium berghei parasite as a surrogate for whole-sporozoite vaccination against P. vivax malaria
npj Vaccines (2022)
-
Advances and opportunities in malaria population genomics
Nature Reviews Genetics (2021)
-
Evolution of the Plasmodium vivax multidrug resistance 1 gene in the Greater Mekong Subregion during malaria elimination
Parasites & Vectors (2020)
-
Genetic diversity of Plasmodium vivax and Plasmodium falciparum lactate dehydrogenases in Myanmar isolates
Malaria Journal (2020)