Abstract
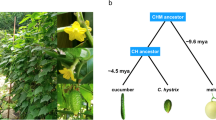
Cucumber is an economically important crop as well as a model system for sex determination studies and plant vascular biology. Here we report the draft genome sequence of Cucumis sativus var. sativus L., assembled using a novel combination of traditional Sanger and next-generation Illumina GA sequencing technologies to obtain 72.2-fold genome coverage. The absence of recent whole-genome duplication, along with the presence of few tandem duplications, explains the small number of genes in the cucumber. Our study establishes that five of the cucumber's seven chromosomes arose from fusions of ten ancestral chromosomes after divergence from Cucumis melo. The sequenced cucumber genome affords insight into traits such as its sex expression, disease resistance, biosynthesis of cucurbitacin and 'fresh green' odor. We also identify 686 gene clusters related to phloem function. The cucumber genome provides a valuable resource for developing elite cultivars and for studying the evolution and function of the plant vascular system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Accessions
NCBI Reference Sequence
References
Tanurdzic, M. & Banks, J.A. Sex-determining mechanisms in land plants. Plant Cell 16, S61–S71 (2004).
Lough, T.J. & Lucas, W.J. Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57, 203–232 (2006).
Xoconostle-Cázares, B. et al. Plant paralog to viral movement protein that potentiates rransport of mRNA into the phloem. Science 283, 94–98 (1999).
Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000).
International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature 436, 793–800 (2005).
Jaillon, O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007).
Ming, R. et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452, 991–996 (2008).
Tuskan, G.A. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 (2006).
Yu, J. et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92 (2002).
Shendure, J., Mitra, R.D., Varma, C. & Church, G.M. Advanced sequencing technologies: methods and goals. Nat. Rev. Genet. 5, 335–344 (2004).
Bentley, D.R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Wang, J. et al. The diploid genome sequence of an Asian individual. Nature 456, 60–65 (2008).
Staub, J.E., Serquen, F.C., Horejsi, T. & Chen, J.-f. Genetic diversity in cucumber (Cucumis sativus L.): IV. An evaluation of Chinese germplasm1. Genet. Resour. Crop Evol. 46, 297–310 (1999).
Arumuganathan, K. & Earle, E. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218 (1991).
Han, Y.H. et al. Distribution of the tandem repeat sequences and karyotyping in cucumber (Cucumis sativus L.) by fluorescence in situ hybridization. Cytogenet. Genome Res. 122, 80–88 (2008).
Ren, Y. et al. An integrated genetic and cytogenetic map of the cucumber genome. PLoS One 4, e5795 (2009).
Bowers, J.E., Chapman, B.A., Rong, J. & Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438 (2003).
Yu, J. et al. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3, e38 (2005).
Fernandez-Silva, I. et al. Bin mapping of genomic and EST-derived SSRs in melon (Cucumis melo L.). Theor. Appl. Genet. 118, 139 (2008).
Schaefer, H., Heibl, C. & Renner, S.S. Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc. Biol. Sci. 276, 843–851 (2009).
Liavonchanka, A. & Feussner, I. Lipoxygenases: occurrence, functions and catalysis. J. Plant Physiol. 163, 348–357 (2006).
Schwab, W., Davidovich-Rikanati, R. & Lewinsohn, E. Biosynthesis of plant-derived flavor compounds. Plant J. 54, 712–732 (2008).
Buescher, R.H. & Buescher, R.W. Production and stability of (E, Z)-2, 6-nonadienal, the major flavor volatile of cucumbers. J. Food Sci. 66, 357–361 (2001).
Cho, M.J., Buescher, R.W., Johnson, M. & Janes, M. Inactivation of pathogenic bacteria by cucumber volatiles (E,Z)-2,6-nonadienal and (E)-2-nonenal. J. Food Prot. 67, 1014–1016 (2004).
Matsui, K. et al. Fatty acid 9- and 13-hydroperoxide lyases from cucumber. FEBS Lett. 481, 183–188 (2000).
Nieto, C. et al. An eIF4E allele confers resistance to an uncapped and non-polyadenylated RNA virus in melon. Plant J. 48, 452–462 (2006).
Wai, T. & Grumet, R. Inheritance of resistance to watermelon mosaic virus in the cucumber line TMG-1: tissue-specific expression and relationship to zucchini yellow mosaic virus resistance. Theor. Appl. Genet. 91, 699–706 (1995).
Taler, D., Galperin, M., Benjamin, I., Cohen, Y. & Kenigsbuch, D. Plant eR genes that encode photorespiratory enzymes confer resistance against disease. Plant Cell 16, 172–184 (2004).
Balkema-Boomstra, A.G. et al. Role of cucurbitacin C in resistance to spider mite Tetranychus urticae in cucumber Cucumis sativus L. J. Chem. Ecol. 29, 225–235 (2003).
Da Costa, C.P. & Jones, C.M. Cucumber beetle resistance and mite susceptibility controlled by the bitter gene in Cucumis sativus L. Science 172, 1145–1146 (1971).
Phillips, D.R., Rasbery, J.M., Bartel, B. & Matsuda, S.P. Biosynthetic diversity in plant triterpene cyclization. Curr. Opin. Plant Biol. 9, 305–314 (2006).
Shibuya, M., Adachi, S. & Ebizuka, Y. Cucurbitadienol synthase, the first committed enzyme for cucurbitacin biosynthesis, is a distinct enzyme from cycloartenol synthase for phytosterol biosynthesis. Tetrahedron 60, 6995–7003 (2004).
Field, B. & Osbourn, A.E. Metabolic diversification–independent assembly of operon-like gene clusters in different plants. Science 320, 543–547 (2008).
Rudich, J., Halevy, A.H. & Kedar, N. Ethylene evolution from cucumber plants as related to sex expression. Plant Physiol. 49, 998–999 (1972).
Pirrung, M.C. Ethylene biosynthesis from 1-aminocyclopropanecarboxylic acid. Acc. Chem. Res. 32, 711–718 (1999).
Stepanova, A.N. & Alonso, J.M. Ethylene signaling pathway. Sci. STKE 2005, cm3 (2005).
Boualem, A. et al. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 321, 836–838 (2008).
Li, Z. et al. Molecular isolation of the M gene suggests that a conserved-residue conversion induces the formation of bisexual flowers in cucumber plants. Genetics 182, 1381–1385 (2009).
Yamagami, T. et al. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 278, 49102–49112 (2003).
Takahashi, H. & Jaffe, M.J. Further studies of auxin and ACC induced feminization in the cucumber plant using ethylene inhibitors. Phyton (Buenos Aires) 44, 81–86 (1984).
DeLong, A., Calderon-Urrea, A. & Dellaporta, S.L. Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 74, 757–768 (1993).
Darwin, C.R. The Movements and Habits of Climbing Plants (Murray, London, 1875).
Boss, P.K. & Thomas, M.R. Association of dwarfism and floral induction with a grape /'green revolution/' mutation. Nature 416, 847–850 (2002).
Galun, E. The cucumber tendril—a new test organ for gibberellic acid. Cell. Mol. Life Sci. 15, 184–185 (1959).
Lange, T. Cloning gibberellin dioxygenase genes from pumpkin endosperm by heterologous expression of enzyme activities in Escherichia coli. Proc. Natl. Acad. Sci. USA 94, 6553–6558 (1997).
Braam, J. In touch: plant responses to mechanical stimuli. New Phytol. 165, 373–389 (2005).
Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 407, 321–326 (2000).
Lin, M.-K., Lee, Y.-J., Lough, T.J., Phinney, B.S. & Lucas, W.J. Analysis of the pumpkin phloem proteome provides insights into angiosperm sieve tube function. Mol. Cell. Proteomics 8, 343–356 (2009).
Rensing, S.A. et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 (2008).
Aki, T., Shigyo, M., Nakano, R., Yoneyama, T. & Yanagisawa, S. Nano scale proteomics revealed the presence of regulatory proteins including rhree FT-Like proteins in phloem and xylem saps from rice. Plant Cell Physiol. 49, 767–790 (2008).
Giavalisco, P., Kapitza, K., Kolasa, A., Buhtz, A. & Kehr, J. Towards the proteome of Brassica napus phloem sap. Proteomics 6, 896–909 (2006).
Dinant, S. et al. Diversity of the superfamily of phloem lectins (phloem protein 2) in angiosperms. Plant Physiol. 131, 114–128 (2003).
Pélissier, H.C., Peters, W.S., Collier, R., van Bel, A.J. & Knoblauch, M. GFP tagging of sieve element occlusion (SEO) proteins results in green fluorescent forisomes. Plant Cell Physiol. 49, 1699–1710 (2008).
Kim, S.J. et al. Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: lessons for database annotations. Phytochemistry 68, 1957–1974 (2007).
Wang, J. et al. RePS: a sequence assembler that masks exact repeats identified from the shotgun data. Genome Res. 12, 824–831 (2002).
Li, R. et al. ReAS: Recovery of ancestral sequences for transposable elements from the unassembled reads of a whole genome shotgun. PLOS Comput. Biol. 1, e43 (2005).
Edgar, R.C. & Myers, E.W. PILER: identification and classification of genomic repeats. Bioinformatics 21 Suppl 1, i152–i158 (2005).
Price, A.L., Jones, N.C. & Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics 21 Suppl 1, i351–i358 (2005).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268 (2007).
Elsik, C.G. et al. Creating a honey bee consensus gene set. Genome Biol. 8, R13 (2007).
Campbell, M.A., Haas, B.J., Hamilton, J.P., Mount, S.M. & Buell, C.R. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics 7, 327 (2006).
Li, H. et al. Test data sets and evaluation of gene prediction programs on the rice genome. J Comp Sci Tech 20, 446–453 (2005).
Majoros, W.H., Pertea, M. & Salzberg, S.L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20, 2878–2879 (2004).
Korf, I. Gene finding in novel genomes. BMC Bioinformatics 5, 59 (2004).
Stanke, M. & Waack, S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 Suppl 2, ii215–ii225 (2003).
Salamov, A.A. & Solovyev, V.V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10, 516–522 (2000).
Birney, E., Clamp, M. & Durbin, R. GeneWise and Genomewise. Genome Res. 14, 988–995 (2004).
Childs, K.L. et al. The TIGR Plant Transcript Assemblies database. Nucleic Acids Res. 35, D846–D851 (2007).
Huang, X., Adams, M.D., Zhou, H. & Kerlavage, A.R. A tool for analyzing and annotating genomic sequences. Genomics 46, 37–45 (1997).
Lowe, T.M. & Eddy, S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Lowe, T.M. & Eddy, S.R. A computational screen for methylation guide snoRNAs in yeast. Science 283, 1168–1171 (1999).
Griffiths-Jones, S. et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33, D121–D124 (2005).
Li, H. et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 34, D572–D580 (2006).
Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Hasegawa, M., Kishino, H. & Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174 (1985).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Crepet, W.L., Nixon, K.C. & Gandolfo, M.A. Fossil evidence and phylogeny: the age of major angiosperm clades based on mesofossil and macrofossil evidence from Cretaceous deposits. Am. J. Botany 91, 1666–1682 (2004).
Wikström, N., Savolainen, V. & Chase, M.W. Evolution of the angiosperms: calibrating the family tree. Proc. Biol. Sci. 268, 2211–2220 (2001).
Acknowledgements
We thank L. Goodman for assistance in editing the manuscript and R. Quatrano, L. Kochian, L. Comai, V. Sundaresan, S. Kamoun and S. Renner for critical readings of the manuscript. This work was funded by the Chinese Ministry of Agriculture (948 program), Ministry of Science and Technology (2006DFA32140, 2007CB815701, 2007CB815703 and 2007CB815705) and Ministry of Finance (1251610601001); the National Natural Science Foundation of China (30871707 and 30725008); the Chinese Academy of Agricultural Sciences (seed grant to S.H.); the Chinese Academy of Science (GJHZ0701-6 and KSCX2-YWN-023); the US Department of Agriculture (National Research Initiative grant 2006-35304-17346 to W.J.L.); the National Science Foundation (grant IOS-07-15513 to W.J.L.); and the Korea Science and Engineering Foundation–Ministry of Education, Science and Technology (WCU R33-10002 and BK21 grants to J.-Y.K.). WKC was partly supported by grants from the Environmental Biotechnology National Core Research Center (R15-2003-012-01003-0) and National Research Laboratory (2009-0066339). This work was also supported by the Shenzhen Municipal and Yantian District Governments and the Society of Entrepreneurs & Ecology. D. Qu and Z. Fang of the Chinese Academy of Agricultural Sciences provided management support for this work.
Author information
Authors and Affiliations
Contributions
S.H., Y.D., Jun Wang and Songgang Li managed the project. S.H., Z.Z., W.J.L., X.G. and R.L. designed the analyses. X.G., H.M., L.L., Yuanyuan Ren, G.T., Y. Lu, Z.X., J.C., A., Z.W., J. Zhang, H. Liang, X.R., M.J., Hailong Yang, R.C., Shifang Liu and X.Z. conducted DNA preparation and sequencing. X.W., B.X., K.L., W.J., Guangcun Li, Z.F., J.S., A.K., E.A.G.v.d.V. and Y.X. contributed new reagents and analytic tools. S.H., Z.Z., W.J.L., X.G., R.L., X.W., B.X., K.L., W.J., J.H., Z.J., Yi Ren, Ying Li, X.L., S.W., Q.S., W.K.C., J.-Y.K., K.H.-U., H.M., Z.C., S.Z., J. Wu, Y.Y., H.K., Y.W., J.G., Y.H., M.L., B. Zhao, Shiqiang Liu, W.F., P.N., H. Zhu, Jun Li, J.R., W.Q., M. Wang, Q.H., B.L., Q.C., Y.B., Z.S., M. Wen, G.Z., Z.Y., Jianwen Li, L.M., H. Liu., Y. Zhou, J. Zhao, X.F., Guoqing Li, L.F., Yingrui Li, D.L., Hancheng Zheng and Shaochuan Li conducted the data analyses. S.H., R.L., Z.Z. and W.J.L. wrote the paper. Y.D., R.S., B. Zhang., S.J., G.Y., S.Y., Hongkun Zheng, Y. Zhang, N.Q., Z.L., L.B., K.K., Huanming Yang and Jian Wang revised the paper.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–20 and Supplementary Tables 1–17, 20. (PDF 2463 kb)
Supplementary Table 18
Phylogenetic relationships of GH20-oxidase genes in cucumber (Csa), Arabidopsis (At), papaya (evm.Tu), poplar (Poptr), grapevine (GSVIVP), rice (BGIOSGA), pumpkin and watermelon. (XLS 440 kb)
Supplementary Table 19
Phylogenetic relationships of expansins in cucumber (Csa), Arabidopsis (At), papaya (evm.Tu), poplar (Poptr) and grapevine (GSVIVP). (XLS 412 kb)
Rights and permissions
About this article
Cite this article
Huang, S., Li, R., Zhang, Z. et al. The genome of the cucumber, Cucumis sativus L.. Nat Genet 41, 1275–1281 (2009). https://doi.org/10.1038/ng.475
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ng.475
This article is cited by
-
Utilization of marker-assisted backcrossing to generate new gynoecious cucumber lines with genetic heritage similar to the indigenous Vietnamese cucumber
Molecular Breeding (2024)
-
Identification of a Novel Candidate Gene for Chilling Tolerance in Pumpkin (Cucurbita moschata) Using Whole-Genome Resequencing
Journal of Plant Biology (2023)
-
Different contributions of bacterial and fungal communities to nitrogen mineralization in Moso bamboo-invaded subtropical forests
Journal of Soils and Sediments (2023)
-
Genome-wide association analysis reveals a novel QTL CsPC1 for pericarp color in cucumber
BMC Genomics (2022)
-
Comprehensive analysis of cucumber C-repeat/dehydration-responsive element binding factor family genes and their potential roles in cold tolerance of cucumber
BMC Plant Biology (2022)