Abstract
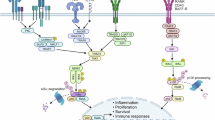
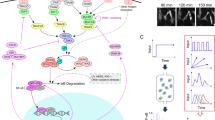
NF-κB transcription factors are critical regulators of immunity, stress responses, apoptosis and differentiation. A variety of stimuli coalesce on NF-κB activation, which can in turn mediate varied transcriptional programs. Consequently, NF-κB-dependent transcription is not only tightly controlled by positive and negative regulatory mechanisms but also closely coordinated with other signaling pathways. This intricate crosstalk is crucial to shaping the diverse biological functions of NF-κB into cell type– and context-specific responses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pahl, H.L. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18, 6853–6866 (1999).
Hayden, M.S. & Ghosh, S. Signaling to NF-κB. Genes Dev. 18, 2195–2224 (2004).
Hayden, M.S. & Ghosh, S. NF-κB in immunobiology. Cell Res. 21, 223–244 (2011).
Pasparakis, M., Luedde, T. & Schmidt-Supprian, M. Dissection of the NF-κB signalling cascade in transgenic and knockout mice. Cell Death Differ. 13, 861–872 (2006).
Gerondakis, S., Grossmann, M., Nakamura, Y., Pohl, T. & Grumont, R. Genetic approaches in mice to understand Rel/NF-κB and IκB function: transgenics and knockouts. Oncogene 18, 6888–6895 (1999).
Courtois, G. & Gilmore, T.D. Mutations in the NF-κB signaling pathway: implications for human disease. Oncogene 25, 6831–6843 (2006).
Karin, M. Nuclear factor-κB in cancer development and progression. Nature 441, 431–436 (2006).
Baker, R.G., Hayden, M.S. & Ghosh, S. NF-κB, inflammation and metabolic disease. Cell Metab. 13, 11–22 (2011).
Kumar, A., Takada, Y., Boriek, A.M. & Aggarwal, B.B. Nuclear factor-κB: its role in health and disease. J. Mol. Med. 82, 434–448 (2004).
Bradley, J.R. & Pober, J.S. Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 20, 6482–6491 (2001).
Chen, Z.J. Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 7, 758–765 (2005).
Tada, K. et al. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-κB activation and protection from cell death. J. Biol. Chem. 276, 36530–36534 (2001).
Liao, G., Zhang, M., Harhaj, E.W. & Sun, S.C. Regulation of the NF-κB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 279, 26243–26250 (2004).
Zarnegar, B.J. et al. Noncanonical NF-κB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 9, 1371–1378 (2008).
Vallabhapurapu, S. et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-κB signaling. Nat. Immunol. 9, 1364–1370 (2008).
Keshet, Y. & Seger, R. The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol. Biol. 661, 3–38 (2010).
Wajant, H., Henkler, F. & Scheurich, P. The TNF-receptor-associated factor family: scaffold molecules for cytokine receptors, kinases and their regulators. Cell Signal. 13, 389–400 (2001).
Chung, J.Y., Park, Y.C., Ye, H. & Wu, H. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 115, 679–688 (2002).
Lee, S.Y. & Choi, Y. TRAF-interacting protein (TRIP): a novel component of the tumor necrosis factor receptor (TNFR)- and CD30-TRAF signaling complexes that inhibits TRAF2-mediated NF-κB activation. J. Exp. Med. 185, 1275–1285 (1997).
Song, H.Y., Rothe, M. & Goeddel, D.V. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-κB activation. Proc. Natl. Acad. Sci. USA 93, 6721–6725 (1996).
Heyninck, K. & Beyaert, R. The cytokine-inducible zinc finger protein A20 inhibits IL-1-induced NF-κB activation at the level of TRAF6. FEBS Lett. 442, 147–150 (1999).
Rothe, M. et al. I-TRAF is a novel TRAF-interacting protein that regulates TRAF-mediated signal transduction. Proc. Natl. Acad. Sci. USA 93, 8241–8246 (1996).
Nomura, F., Kawai, T., Nakanishi, K. & Akira, S. NF-κB activation through IKK-i-dependent I-TRAF/TANK phosphorylation. Genes Cells 5, 191–202 (2000).
Hacker, H. & Karin, M. Regulation and function of IKK and IKK-related kinases. Sci. STKE 2006, re13 (2006).
Oganesyan, G. et al. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439, 208–211 (2006).
Saha, S.K. et al. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 25, 3257–3263 (2006).
Kawai, T. et al. Interferon-α induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 5, 1061–1068 (2004).
Zeng, W. et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141, 315–330 (2010).
West, A.P. et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472, 476–480 (2011).
Kopp, E. et al. ECSIT is an evolutionarily conserved intermediate in the Toll/IL-1 signal transduction pathway. Genes Dev. 13, 2059–2071 (1999).
Vogel, R.O. et al. Cytosolic signaling protein Ecsit also localizes to mitochondria where it interacts with chaperone NDUFAF1 and functions in complex I assembly. Genes Dev. 21, 615–624 (2007).
Wong, B.R. et al. TRANCE, a TNF family member, activates Akt/PKB through a signaling complex involving TRAF6 and c-Src. Mol. Cell 4, 1041–1049 (1999).
Lomaga, M.A. et al. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 13, 1015–1024 (1999).
Bryce, P.J., Oyoshi, M.K., Kawamoto, S., Oettgen, H.C. & Tsitsikov, E.N. TRAF1 regulates Th2 differentiation, allergic inflammation and nuclear localization of the Th2 transcription factor, NIP45. Int. Immunol. 18, 101–111 (2006).
Lieberson, R. et al. Tumor necrosis factor receptor-associated factor (TRAF)2 represses the T helper cell type 2 response through interaction with NFAT-interacting protein (NIP45). J. Exp. Med. 194, 89–98 (2001).
Lamothe, B. et al. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of IκB kinase activation. J. Biol. Chem. 282, 4102–4112 (2007).
Liang, J. et al. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-κB signaling. J. Exp. Med. 207, 2959–2973 (2010).
Thomas, G.S., Zhang, L., Blackwell, K. & Habelhah, H. Phosphorylation of TRAF2 within its RING domain inhibits stress-induced cell death by promoting IKK and suppressing JNK activation. Cancer Res. 69, 3665–3672 (2009).
Zhang, L., Blackwell, K., Altaeva, A., Shi, Z. & Habelhah, H. TRAF2 phosphorylation promotes NF-κB-dependent gene expression and inhibits oxidative stress-induced cell death. Mol. Biol. Cell 22, 128–140 (2011).
Li, S., Wang, L. & Dorf, M.E. PKC phosphorylation of TRAF2 mediates IKKα/β recruitment and K63-linked polyubiquitination. Mol. Cell 33, 30–42 (2009).
Meylan, E. & Tschopp, J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem. Sci. 30, 151–159 (2005).
Liu, Z.G. Molecular mechanism of TNF signaling and beyond. Cell Res. 15, 24–27 (2005).
Ea, C.K., Deng, L., Xia, Z.P., Pineda, G. & Chen, Z.J. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22, 245–257 (2006).
Gerlach, B. et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 (2011).
Micheau, O. & Tschopp, J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114, 181–190 (2003).
Jones, S.J. et al. TNF recruits TRADD to the plasma membrane but not the trans-Golgi network, the principal subcellular location of TNF-R1. J. Immunol. 162, 1042–1048 (1999).
Holler, N. et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat. Immunol. 1, 489–495 (2000).
Degterev, A. et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 4, 313–321 (2008).
Cho, Y.S. et al. Phosphorylation-driven assembly of the RIP1–RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123 (2009).
Vandenabeele, P., Declercq, W., Van Herreweghe, F. & Vanden Berghe, T. The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci. Signal. 3, re4 (2010).
Vandenabeele, P., Declercq, W. & Vanden Berghe, T. Necrotic cell death and 'necrostatins': now we can control cellular explosion. Trends Biochem. Sci. 33, 352–355 (2008).
Benedict, C.A., Norris, P.S. & Ware, C.F. To kill or be killed: viral evasion of apoptosis. Nat. Immunol. 3, 1013–1018 (2002).
Lin, Y., Devin, A., Rodriguez, Y. & Liu, Z.G. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 13, 2514–2526 (1999).
Hitomi, J. et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135, 1311–1323 (2008).
Wang, L., Du, F. & Wang, X. TNF-α induces two distinct caspase-8 activation pathways. Cell 133, 693–703 (2008).
Knox, P.G., Davies, C.C., Ioannou, M. & Eliopoulos, A.G. The death domain kinase RIP1 links the immunoregulatory CD40 receptor to apoptotic signaling in carcinomas. J. Cell Biol. 192, 391–399 (2011).
Moquin, D. & Chan, F.K. The molecular regulation of programmed necrotic cell injury. Trends Biochem. Sci. 35, 434–441 (2010).
O'Donnell, M.A., Legarda-Addison, D., Skountzos, P., Yeh, W.C. & Ting, A.T. Ubiquitination of RIP1 regulates an NF-κB-independent cell-death switch in TNF signaling. Curr. Biol. 17, 418–424 (2007).
Ramnarain, D.B. et al. RIP1 links inflammatory and growth factor signaling pathways by regulating expression of the EGFR. Cell Death Differ. 15, 344–353 (2008).
Habib, A.A. et al. The epidermal growth factor receptor engages receptor interacting protein and nuclear factor-κB (NF-κB)-inducing kinase to activate NF-κB. Identification of a novel receptor-tyrosine kinase signalosome. J. Biol. Chem. 276, 8865–8874 (2001).
Park, S. et al. RIP1 activates PI3K-Akt via a dual mechanism involving NF-κB-mediated inhibition of the mTOR-S6K–IRS1 negative feedback loop and down-regulation of PTEN. Cancer Res. 69, 4107–4111 (2009).
Ikeda, F. et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471, 637–641 (2011).
Tokunaga, F. et al. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636 (2011).
Scheidereit, C. IκB kinase complexes: gateways to NF-κB activation and transcription. Oncogene 25, 6685–6705 (2006).
Ozes, O.N. et al. NF-κB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature 401, 82–85 (1999).
Agarwal, A. et al. The AKT/IκB kinase pathway promotes angiogenic/metastatic gene expression in colorectal cancer by activating nuclear factor-κB and β-catenin. Oncogene 24, 1021–1031 (2005).
Dan, H.C. et al. Akt-dependent regulation of NF-κB is controlled by mTOR and Raptor in association with IKK. Genes Dev. 22, 1490–1500 (2008).
Ghosh, S. et al. Essential role of tuberous sclerosis genes TSC1 and TSC2 in NF-κB activation and cell survival. Cancer Cell 10, 215–226 (2006).
May, M.J. et al. Selective inhibition of NF-κB activation by a peptide that blocks the interaction of NEMO with the IκB kinase complex. Science 289, 1550–1554 (2000).
Schomer-Miller, B., Higashimoto, T., Lee, Y.K. & Zandi, E. Regulation of IκB kinase (IKK) complex by IKKγ-dependent phosphorylation of the T-loop and C terminus of IKKβ. J. Biol. Chem. 281, 15268–15276 (2006).
Comer, F.I. & Hart, G.W. O-Glycosylation of nuclear and cytosolic proteins. Dynamic interplay between O-GlcNAc and O-phosphate. J. Biol. Chem. 275, 29179–29182 (2000).
Kawauchi, K., Araki, K., Tobiume, K. & Tanaka, N. Loss of p53 enhances catalytic activity of IKKβ through O-linked β-N-acetyl glucosamine modification. Proc. Natl. Acad. Sci. USA 106, 3431–3436 (2009).
Frelin, C. et al. Inhibition of the NF-κB survival pathway via caspase-dependent cleavage of the IKK complex scaffold protein and NF-κB essential modulator NEMO. Cell Death Differ. 15, 152–160 (2008).
Tang, G., Yang, J., Minemoto, Y. & Lin, A. Blocking caspase-3-mediated proteolysis of IKKβ suppresses TNF-α-induced apoptosis. Mol. Cell 8, 1005–1016 (2001).
Levkau, B., Scatena, M., Giachelli, C.M., Ross, R. & Raines, E.W. Apoptosis overrides survival signals through a caspase-mediated dominant-negative NF-κB loop. Nat. Cell Biol. 1, 227–233 (1999).
Reuther, J.Y. & Baldwin, A.S. Jr. Apoptosis promotes a caspase-induced amino-terminal truncation of IκBα that functions as a stable inhibitor of NF-κB. J. Biol. Chem. 274, 20664–20670 (1999).
Vilimas, T. et al. Targeting the NF-κB signaling pathway in Notch1-induced T-cell leukemia. Nat. Med. 13, 70–77 (2007).
Song, L.L. et al. Notch-1 associates with IKKα and regulates IKK activity in cervical cancer cells. Oncogene 27, 5833–5844 (2008).
Osipo, C., Golde, T.E., Osborne, B.A. & Miele, L.A. Off the beaten pathway: the complex cross talk between Notch and NF-κB. Lab. Invest. 88, 11–17 (2008).
Hinz, M. & Scheidereit, C. Striking back at the activator: how IκB kinase terminates antigen receptor responses. Sci. STKE 2007, pe19 (2007).
Perkins, N.D. Integrating cell-signalling pathways with NF-κB and IKK function. Nat. Rev. Mol. Cell Biol. 8, 49–62 (2007).
Chariot, A. The NF-κB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol. 19, 404–413 (2009).
Hu, M.C. et al. IκB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell 117, 225–237 (2004).
Chapuis, N. et al. IκB kinase overcomes PI3K/Akt and ERK/MAPK to control FOXO3a activity in acute myeloid leukemia. Blood 116, 4240–4250 (2010).
Lee, D.F. et al. IKKβ suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell 130, 440–455 (2007).
Suzuki, K. & Verma, I.M. Phosphorylation of SNAP-23 by IκB kinase 2 regulates mast cell degranulation. Cell 134, 485–495 (2008).
Guo, Z., Turner, C. & Castle, D. Relocation of the t-SNARE SNAP-23 from lamellipodia-like cell surface projections regulates compound exocytosis in mast cells. Cell 94, 537–548 (1998).
Hepp, R. et al. Phosphorylation of SNAP-23 regulates exocytosis from mast cells. J. Biol. Chem. 280, 6610–6620 (2005).
Polgar, J., Lane, W.S., Chung, S.H., Houng, A.K. & Reed, G.L. Phosphorylation of SNAP-23 in activated human platelets. J. Biol. Chem. 278, 44369–44376 (2003).
Waterfield, M.R., Zhang, M., Norman, L.P. & Sun, S.C. NF-κB1/p105 regulates lipopolysaccharide-stimulated MAP kinase signaling by governing the stability and function of the Tpl2 kinase. Mol. Cell 11, 685–694 (2003).
Beinke, S. et al. NF-κB1 p105 negatively regulates TPL-2 MEK kinase activity. Mol. Cell Biol. 23, 4739–4752 (2003).
Beinke, S., Robinson, M.J., Hugunin, M. & Ley, S.C. Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IκB kinase-induced proteolysis of NF-κB1 p105. Mol. Cell. Biol. 24, 9658–9667 (2004).
Waterfield, M., Jin, W., Reiley, W., Zhang, M. & Sun, S.C. IκB kinase is an essential component of the Tpl2 signaling pathway. Mol. Cell Biol. 24, 6040–6048 (2004).
Bouwmeester, T. et al. A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat. Cell Biol. 6, 97–105 (2004).
Ferrier, R. et al. Physical interaction of the bHLH LYL1 protein and NF-κB1 p105. Oncogene 18, 995–1005 (1999).
Li, Z., Zhang, J., Chen, D. & Shu, H.B. Casper/c-FLIP is physically and functionally associated with NF-κB1 p105. Biochem. Biophys. Res. Commun. 309, 980–985 (2003).
Lang, V. et al. ABIN-2 forms a ternary complex with TPL-2 and NF-κB1 p105 and is essential for TPL-2 protein stability. Mol. Cell Biol. 24, 5235–5248 (2004).
Verstrepen, L., Carpentier, I., Verhelst, K. & Beyaert, R. ABINs: A20 binding inhibitors of NF-κB and apoptosis signaling. Biochem. Pharmacol. 78, 105–114 (2009).
Papoutsopoulou, S. et al. ABIN-2 is required for optimal activation of Erk MAP kinase in innate immune responses. Nat. Immunol. 7, 606–615 (2006).
Oshima, S. et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature 457, 906–909 (2009).
Lee, S. et al. IκB kinase β phosphorylates Dok1 serines in response to TNF, IL-1, or gamma radiation. Proc. Natl. Acad. Sci. USA 101, 17416–17421 (2004).
Clark, K. et al. Novel cross-talk within the IKK family controls innate immunity. Biochem. J. 27, 93–104 (2011).
Kawagoe, T. et al. TANK is a negative regulator of Toll-like receptor signaling and is critical for the prevention of autoimmune nephritis. Nat. Immunol. 10, 965–972 (2009).
Barbie, D.A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009).
Boehm, J.S. et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129, 1065–1079 (2007).
Shen, R.R. & Hahn, W.C. Emerging roles for the non-canonical IKKs in cancer. Oncogene 30, 631–641 (2011).
Tilg, H. & Moschen, A.R. Inflammatory mechanisms in the regulation of insulin resistance. Mol. Med. 14, 222–231 (2008).
Gao, Z. et al. Serine phosphorylation of insulin receptor substrate 1 by inhibitor κB kinase complex. J. Biol. Chem. 277, 48115–48121 (2002).
He, J. et al. Interleukin-1α inhibits insulin signaling with phosphorylating insulin receptor substrate-1 on serine residues in 3T3–L1 adipocytes. Mol. Endocrinol. 20, 114–124 (2006).
Nakamori, Y. et al. Myosin motor Myo1c and its receptor NEMO/IKK-gamma promote TNF-α-induced serine307 phosphorylation of IRS-1. J. Cell Biol. 173, 665–671 (2006).
Yaspelkis, B.B. III, Kvasha, I.A. & Figueroa, T.Y. High-fat feeding increases insulin receptor and IRS-1 coimmunoprecipitation with SOCS-3, IKKα/β phosphorylation and decreases PI-3 kinase activity in muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 296, R1709–R1715 (2009).
Rao, T.P. & Kuhl, M. An updated overview on Wnt signaling pathways: a prelude for more. Circ. Res. 106, 1798–1806 (2010).
Albanese, C. et al. IKKα regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol. Biol. Cell 14, 585–599 (2003).
Carayol, N. & Wang, C.Y. IKKα stabilizes cytosolic β-catenin by inhibiting both canonical and non-canonical degradation pathways. Cell Signal. 18, 1941–1946 (2006).
Lamberti, C. et al. Regulation of β-catenin function by the IκB kinases. J. Biol. Chem. 276, 42276–42286 (2001).
Park, K.J., Krishnan, V., O'Malley, B.W., Yamamoto, Y. & Gaynor, R.B. Formation of an IKKα-dependent transcription complex is required for estrogen receptor-mediated gene activation. Mol. Cell 18, 71–82 (2005).
Wu, R.C. et al. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) Coactivator activity by IκB kinase. Mol. Cell Biol. 22, 3549–3561 (2002).
Kwak, Y.T. et al. IκB kinase α regulates subcellular distribution and turnover of cyclin D1 by phosphorylation. J. Biol. Chem. 280, 33945–33952 (2005).
Song, L. et al. A novel role of IKKα in the mediation of UVB-induced G0/G1 cell cycle arrest response by suppressing Cyclin D1 expression. Biochim. Biophys. Acta. 1803, 323–332 (2010).
Kwak, Y.T. et al. Cells lacking IKKα show nuclear cyclin D1 overexpression and a neoplastic phenotype: role of IKKα as a tumor suppressor. Mol. Cancer Res. 9, 341–349 (2011).
Hoshino, K. et al. IκB kinase-α is critical for interferon-α production induced by Toll-like receptors 7 and 9. Nature 440, 949–953 (2006).
Wang, R.P. et al. Differential regulation of IKK α-mediated activation of IRF3/7 by NIK. Mol. Immunol. 45, 1926–1934 (2008).
Balkhi, M.Y., Fitzgerald, K.A. & Pitha, P.M. IKKα negatively regulates IRF-5 function in a MyD88-TRAF6 pathway. Cell. Signal. 22, 117–127 (2010).
Anest, V. et al. A nucleosomal function for IκB kinase-α in NF-κB-dependent gene expression. Nature 423, 659–663 (2003).
Yamamoto, Y., Verma, U.N., Prajapati, S., Kwak, Y.T. & Gaynor, R.B. Histone H3 phosphorylation by IKK-α is critical for cytokine-induced gene expression. Nature 423, 655–659 (2003).
Anest, V., Cogswell, P.C. & Baldwin, A.S. Jr. IκB kinase α and p65/RelA contribute to optimal epidermal growth factor-induced c-fos gene expression independent of IκBα degradation. J. Biol. Chem. 279, 31183–31189 (2004).
Saccani, S., Pantano, S. & Natoli, G. p38-Dependent marking of inflammatory genes for increased NF-κB recruitment. Nat. Immunol. 3, 69–75 (2002).
Li, Q. et al. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes Dev. 13, 1322–1328 (1999).
Takeda, K. et al. Limb and skin abnormalities in mice lacking IKKα. Science 284, 313–316 (1999).
Hu, Y. et al. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKα subunit of IκB kinase. Science 284, 316–320 (1999).
Sil, A.K., Maeda, S., Sano, Y., Roop, D.R. & Karin, M. IκB kinase-α acts in the epidermis to control skeletal and craniofacial morphogenesis. Nature 428, 660–664 |(2004).
Lawrence, T., Bebien, M., Liu, G.Y., Nizet, V. & Karin, M. IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature 434, 1138–1143 (2005).
Zhu, F. et al. IKKα shields 14–3-3sigma, a G(2)/M cell cycle checkpoint gene, from hypermethylation, preventing its silencing. Mol. Cell 27, 214–227 (2007).
Liu, B. et al. IKKα is required to maintain skin homeostasis and prevent skin cancer. Cancer Cell 14, 212–225 (2008).
Hu, Y. et al. IKKα controls formation of the epidermis independently of NF-κB. Nature 410, 710–714 (2001).
Ohazama, A. et al. A dual role for IKKα in tooth development. Dev. Cell 6, 219–227 (2004).
Descargues, P. et al. IKKα is a critical coregulator of a Smad4-independent TGFβ-Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc. Natl. Acad. Sci. USA 105, 2487–2492 (2008).
Ravi, R. et al. p53-mediated repression of nuclear factor-κB RelA via the transcriptional integrator p300. Cancer Res. 58, 4531–4536 (1998).
Webster, G.A. & Perkins, N.D. Transcriptional cross talk between NF-κB and p53. Mol. Cell Biol. 19, 3485–3495 (1999).
Huang, W.C., Ju, T.K., Hung, M.C. & Chen, C.C. Phosphorylation of CBP by IKKα promotes cell growth by switching the binding preference of CBP from p53 to NF-κB. Mol. Cell 26, 75–87 (2007).
Luo, J.L. et al. Nuclear cytokine-activated IKKα controls prostate cancer metastasis by repressing Maspin. Nature 446, 690–694 (2007).
Bracken, C.P., Whitelaw, M.L. & Peet, D.J. Activity of hypoxia-inducible factor 2α is regulated by association with the NF-κB essential modulator. J. Biol. Chem. 280, 14240–14251 (2005).
Cockman, M.E. et al. Posttranslational hydroxylation of ankyrin repeats in IκB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH). Proc. Natl. Acad. Sci. USA 103, 14767–14772 (2006).
Bettermann, K. et al. TAK1 suppresses a NEMO-dependent but NF-κB-independent pathway to liver cancer. Cancer Cell 17, 481–496 (2010).
Biton, S. & Ashkenazi, A. NEMO and RIP1 control cell fate in response to extensive DNA damage via TNF-α feedforward signaling. Cell 145, 92–103 (2011).
Matsuzawa, A. et al. Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science 321, 663–668 (2008).
Zhao, T. et al. The NEMO adaptor bridges the nuclear factor-κB and interferon regulatory factor signaling pathways. Nat. Immunol. 8, 592–600 (2007).
Chen, L.F. & Greene, W.C. Shaping the nuclear action of NF-κB. Nat. Rev. Mol. Cell Biol. 5, 392–401 (2004).
Zhong, H., SuYang, H., Erdjument-Bromage, H., Tempst, P. & Ghosh, S. The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell 89, 413–424 (1997).
Zhong, H., May, M.J., Jimi, E. & Ghosh, S. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell 9, 625–636 (2002).
Vermeulen, L., De Wilde, G., Van Damme, P., Vanden Berghe, W. & Haegeman, G. Transcriptional activation of the NF-κB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1). EMBO J. 22, 1313–1324 (2003).
Leitges, M. et al. Targeted disruption of the ζPKC gene results in the impairment of the NF-κB pathway. Mol. Cell 8, 771–780 (2001).
Schwabe, R.F. & Brenner, D.A. Role of glycogen synthase kinase-3 in TNF-α-induced NF-κB activation and apoptosis in hepatocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 283, G204–G211 (2002).
Wang, D., Westerheide, S.D., Hanson, J.L. & Baldwin, A.S. Jr. Tumor necrosis factor α-induced phosphorylation of RelA/p65 on Ser529 is controlled by casein kinase II. J. Biol. Chem. 275, 32592–32597 (2000).
Sakurai, H., Chiba, H., Miyoshi, H., Sugita, T. & Toriumi, W. IκB kinases phosphorylate NF-κB p65 subunit on serine 536 in the transactivation domain. J. Biol. Chem. 274, 30353–30356 (1999).
Bohuslav, J., Chen, L.F., Kwon, H., Mu, Y. & Greene, W.C. p53 induces NF-κB activation by an IκB kinase-independent mechanism involving phosphorylation of p65 by ribosomal S6 kinase 1. J. Biol. Chem. 279, 26115–26125 (2004).
Fujita, F. et al. Identification of NAP1, a regulatory subunit of IκB kinase-related kinases that potentiates NF-κB signaling. Mol. Cell Biol. 23, 7780–7793 (2003).
Haller, D., Russo, M.P., Sartor, R.B. & Jobin, C. IKKβ and phosphatidylinositol 3-kinase/Akt participate in non-pathogenic Gram-negative enteric bacteria-induced RelA phosphorylation and NF-κB activation in both primary and intestinal epithelial cell lines. J. Biol. Chem. 277, 38168–38178 (2002).
Rocha, S., Garrett, M.D., Campbell, K.J., Schumm, K. & Perkins, N.D. Regulation of NF-κB and p53 through activation of ATR and Chk1 by the ARF tumour suppressor. EMBO J. 24, 1157–1169 (2005).
Chen, L., Fischle, W., Verdin, E. & Greene, W.C. Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293, 1653–1657 (2001).
Chen, L.F., Mu, Y. & Greene, W.C. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 21, 6539–6548 (2002).
Kiernan, R. et al. Post-activation turn-off of NF-κB-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 278, 2758–2766 (2003).
Ea, C.K. & Baltimore, D. Regulation of NF-κB activity through lysine monomethylation of p65. Proc. Natl. Acad. Sci. USA 106, 18972–18977 (2009).
Li, Y. et al. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-κB-dependent inflammatory genes. Relevance to diabetes and inflammation. J. Biol. Chem. 283, 26771–26781 (2008).
Yang, X.D. et al. Negative regulation of NF-κB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 28, 1055–1066 (2009).
Lu, T. et al. Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl. Acad. Sci. USA 107, 46–51 (2010).
Wietek, C., Miggin, S.M., Jefferies, C.A. & O'Neill, L.A. Interferon regulatory factor-3-mediated activation of the interferon-sensitive response element by Toll-like receptor (TLR) 4 but not TLR3 requires the p65 subunit of NF-κ. J. Biol. Chem. 278, 50923–50931 (2003).
Taniguchi, T., Ogasawara, K., Takaoka, A. & Tanaka, N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19, 623–655 (2001).
Ogawa, S. et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell 122, 707–721 (2005).
Apostolou, E. & Thanos, D. Virus Infection Induces NF-κB-dependent interchromosomal associations mediating monoallelic IFN-β gene expression. Cell 134, 85–96 (2008).
Cheng, C.S. et al. The specificity of innate immune responses is enforced by repression of interferon response elements by NF-κB p50. Sci. Signal. 4, ra11 (2011).
Wei, L. et al. NFκB negatively regulates interferon-induced gene expression and anti-influenza activity. J. Biol. Chem. 281, 11678–11684 (2006).
Sha, W.C., Liou, H.C., Tuomanen, E.I. & Baltimore, D. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell 80, 321–330 (1995).
Perkins, N.D. Achieving transcriptional specificity with NF-κB. Int. J. Biochem. Cell Biol. 29, 1433–1448 (1997).
Sanceau, J., Kaisho, T., Hirano, T. & Wietzerbin, J. Triggering of the human interleukin-6 gene by interferon-gamma and tumor necrosis factor-α in monocytic cells involves cooperation between interferon regulatory factor-1, NFκB, and Sp1 transcription factors. J. Biol. Chem. 270, 27920–27931 (1995).
Wang, T., Lafuse, W.P. & Zwilling, B.S. NFκB and Sp1 elements are necessary for maximal transcription of toll-like receptor 2 induced by Mycobacterium avium. J. Immunol. 167, 6924–6932 (2001).
Stein, B. et al. Cross-coupling of the NF-κB p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J. 12, 3879–3891 (1993).
Krappmann, D. et al. The IκB kinase complex and NF-κB act as master regulators of lipopolysaccharide-induced gene expression and control subordinate activation of AP-1. Mol. Cell Biol. 24, 6488–6500 (2004).
Fujioka, S. et al. NF-κB and AP-1 connection: mechanism of NF-κB-dependent regulation of AP-1 activity. Mol. Cell. Biol. 24, 7806–7819 (2004).
Koga, K. et al. Cyclic adenosine monophosphate suppresses the transcription of proinflammatory cytokines via the phosphorylated c-Fos protein. Immunity 30, 372–383 (2009).
Stein, B. & Baldwin, A.S. Jr. Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-κB. Mol. Cell Biol. 13, 7191–7198 (1993).
Xia, C., Cheshire, J.K., Patel, H. & Woo, P. Cross-talk between transcription factors NF-κB and C/EBP in the transcriptional regulation of genes. Int. J. Biochem. Cell Biol. 29, 1525–1539 (1997).
Papa, S., Zazzeroni, F., Pham, C.G., Bubici, C. & Franzoso, G. Linking JNK signaling to NF-κB: a key to survival. J. Cell Sci. 117, 5197–5208 (2004).
Tergaonkar, V., Pando, M., Vafa, O., Wahl, G. & Verma, I. p53 stabilization is decreased upon NFκB activation: a role for NFκB in acquisition of resistance to chemotherapy. Cancer Cell 1, 493–503 (2002).
Rius, J. et al. NF-κB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1α. Nature 453, 807–811 (2008).
Greten, F.R. et al. NF-κB is a negative regulator of IL-1β secretion as revealed by genetic and pharmacological inhibition of IKKβ. Cell 130, 918–931 (2007).
Acknowledgements
Supported by the US National Institutes of Health (R37-AI33443) and the American Heart Association (A.O.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Oeckinghaus, A., Hayden, M. & Ghosh, S. Crosstalk in NF-κB signaling pathways. Nat Immunol 12, 695–708 (2011). https://doi.org/10.1038/ni.2065
Published:
Issue date:
DOI: https://doi.org/10.1038/ni.2065
This article is cited by
-
CLK2 mediates IκBα-independent early termination of NF-κB activation by inducing cytoplasmic redistribution and degradation
Nature Communications (2024)
-
Impact of modeled microgravity stress on innate immunity in a beneficial animal-microbe symbiosis
Scientific Reports (2024)
-
hUC-MSCs-derived MFGE8 ameliorates locomotor dysfunction via inhibition of ITGB3/ NF-κB signaling in an NMO mouse model
npj Regenerative Medicine (2024)
-
Ischemia-reperfusion injury: molecular mechanisms and therapeutic targets
Signal Transduction and Targeted Therapy (2024)
-
Liensinine reduces acute lung injury brought on by lipopolysaccharide by inhibiting the activation of the NF-κB signaling pathway through modification of the Src/TRAF6/TAK1 axis
Inflammopharmacology (2024)