Abstract
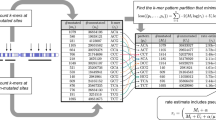
Understanding of the mechanism of DNA mutation process is critical for studying the functional consequences of genetic variation in clinical medicine (eg. drug response) as well as other complex traits (eg. gene expression and cause of common diseases). During the last several decades, many probabilistic models of DNA nucleotide substitution have been proposed for studying this process. A common feature of these mutation models is that they assume the nucleotides evolve independently at each site. In other words, they are sequence context-independent models. However, based on various biochemical studies, it is now recognized that the DNA mutation process resulting in substitutions in both coding and non-coding regions may depend on sequence context. We proposed here a more realistic sequence context-dependent mutation model, which could contribute to the better understanding of the DNA mutation spectrum in genomes.We also showed its application to protein evolution by separating the mutation bias and selection bias in amino acid substitutions.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, W. Estimation of DNA Sequence Context-dependent Mutation Rates Using Primate Genomic Sequences: Application to Estimation of Selection Bias in Protein (Human TP53) Evolution. Nat Prec (2007). https://doi.org/10.1038/npre.2007.1243.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2007.1243.1