Abstract
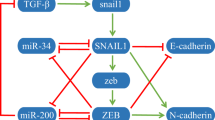
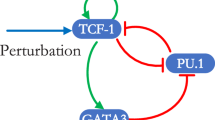
Several meso-scale biological intracellular regulatory networks that have specified directionality of interactions have been recently assembled from experimental literature. Directed networks where links are characterized as positive or negative can be converted to systems of differential equations and analyzed as dynamical systems. Such analyses have shown that networks containing only sign-consistent loops, such as positive feed-forward and feedback loops function as monotone systems that display well-ordered behavior. Perturbations to monotone systems have unambiguous global effects and a predictability characteristic that confers advantages for robustness and adaptability. We find that three intracellular regulatory networks: bacterial and yeast transcriptional networks and a mammalian signaling network contain far more sign-consistent feedback and feed-forward loops than expected for shuffled networks. Inconsistent loops with negative links can be more easily removed from real regulatory networks as compared to shuffled networks. This topological feature in real networks emerges from the presence of hubs that are enriched for either negative or positive links, and is not due to a preference for double negative links in paths. These observations indicate that intracellular regulatory networks may be close to monotone systems and that this network topology contributes to the dynamic stability.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ma'ayan, A., Iyengar, R. & Sontag, E. Intracellular Regulatory Networks are close to Monotone Systems. Nat Prec (2007). https://doi.org/10.1038/npre.2007.25.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2007.25.1