Abstract
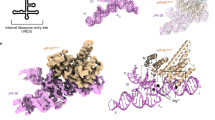
Unlike prokaryotes, a specialized eukaryotic initiation factor 2 (eIF2), in the form of the ternary complex eIF2*GTP*Met-tRNAiMet is utilized to deliver the initiator tRNA to the ribosome within all eukaryotic cells1. Phosphorylation of eIF2 is known to be central to the global regulation of protein synthesis under stress conditions and infection2. Another distinctive feature of eukaryotic translation is scanning of mRNA 5'-leaders, whose origin in evolution may be relevant to the appearance of eIF2 in eukaryotes. Translation initiation on the hepatitis C virus (HCV) internal ribosome entry site (IRES) occurs without scanning3,4. Whether these unique features of the HCV IRES account for the formation of the final 80S initiation complex is unknown. Here we show that the HCV IRES-directed translation can occur without either eIF2 or its GTPase activating protein eIF5. In addition to the general eIF2- and eIF5-dependent pathway of 80S complex assembly, the HCV IRES makes use of a prokaryotic-like pathway which involves eIF5B, the analogue of bacterial IF25,6, instead of eIF2. This switch from a eukaryotic-like mode of AUG selection to a "bacterial" one occurs when eIF2 is inactivated by phosphorylation, a way with which host cells counteract infection. The relative resistance of HCV IRES-directed translation to eIF2 phosphorylation may represent one more line of defense used by this virus against host antiviral responses and can contribute to the well known resistance of HCV to interferon based therapy.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shatsky, I., Terenin, I., Dmitriev, S. et al. Eukaryotic translation initiation machinery can operate in a prokaryotic-like mode without eIF2. Nat Prec (2008). https://doi.org/10.1038/npre.2008.1518.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2008.1518.1