Abstract
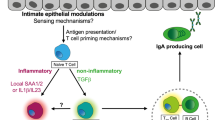
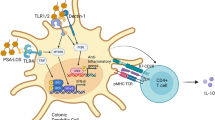
Mucosal immunity distinguishes not only different microbial antigens but also separates those of pathogens from those of commensals. How this is done is unknown. The present view is that the pathogen/commensal determination of antigens depends upon as yet to be discovered molecular patterns. Here I review the biological feasibility that it also involves the detection of the invasive differences in their motility towards the gut wall when they are sampled by differently biased methods. By their nature, pathogens and commensals have different motility – invasive and noninvasive – in regard to the epithelium. The immune system is in a position to detect such motility differences. This biological opportunity arises since different microbe sampling methods can “catch” different groups of microbes depending upon how their motility interacts with the epithelium. A biological method biased to sample those with invasive motility—pathogens—could be achieved by ‘honey pot traps’ that preferentially (but not exclusively) sample microbes that have a taxis to breaches in the epithelium. A biological method biased to sample those that are noninvasive—commensals—could be done by capturing microbes that are passively and stably residing in the biofilm “offshore” of the epithelium. Such differential sampling strategies would seem to relate to those carried out respectively by (i) M-cells (working with subepithelial dome dendritic cells), and (ii) sub- and intraepithelial dendritic cells.The interactions of antigen presentation can be arranged so that the immune system links antigens from biased microbial sampling with pathogenic or commensal appropriate immune responses. Such immune classification could feasibly occur biologically through a winner-take-all competition between inhibiting and activating antigen presentation. Winner-takes-all types of processing classification are already known to underlie the biologically interactions between neurons that classify sensory inputs making it also plausible that they are exploited by the immune system. In pathogen identification, M-cell antigens would be activating and biofilm antigens inhibitory, and vise versa for commensal identification. This winner-take-all competition between antigen presentation would act to amplify small statistical biases in the two samples linked to invasiveness/noninvasiveness into a reliable pathogen/commensal distinction. This process would both complement, and acts as independent guarantor, upon the alternative pathogenicity/commensality recognition provided by molecular pattern recognition.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Skoyles, J. How gut sampling and microbial invasiveness/noninvasiveness provides mucosal immunity with a nonmolecular pattern means to distinguish commensals from pathogens: A review. Nat Prec (2008). https://doi.org/10.1038/npre.2008.1895.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2008.1895.1