Abstract
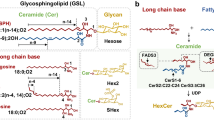
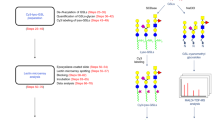
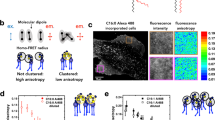
Glycosphingolipids (GSLs) synthesized in Golgi apparatus by sequential transfer of sugar residues to a ceramide lipid anchor are ubiquitously distributing on vertebrate plasma membranes. Standardized method allowing for high throughput structural profiling and functional characterization of living cell surface GSLs is of growing importance because they function as crucial signal transduction molecules in various processes of dynamic cellular recognitions. However, methods are not available for amplification of GSLs, while the genomic scale PCR amplification permits large-scale mammalian proteomic analysis.&x3000;Here we communicate such an approach to a novel "omics", namely glycosphingolipidomics based on the glycoblotting method. The method, which involves selective ozonolysis of the C-C double bond in ceramide moiety and subsequent enrichment of generated GSL-aldehydes by chemical ligation using aminooxy-functionalized gold nanoparticle (aoGNP) should be of widespread utility for identifying and characterizing whole GSLs present in the living cell surfaces. The present protocol using glycoblotting permitted MALDI-TOFMS-based high throughput structural profiling of mouse brain gangliosides such as GM1, GD1a/GD1b, and GT1b for adult or GD3 in case for embryonic mouse. When mouse melanoma B16 cells were subjected to this protocol, it was demonstrated that gangliosides enriched from the plasma membranes are only GM3 bearing microheteogeneity in the structure of N-acyl chain. Surface plasmon resonance analysis revealed that aoGNP displaying whole GSLs blotted from mouse B16 melanoma cell surfaces can be used directly for monitoring specific interaction with self-assembled monolayer (SAM) of Gg3Cer (gangliotriaosylceramide). Our results indicate that GSL-selective enrichment onto aoGNP from living cell surfaces allows for rapid reconstruction of plasma membrane models mimicking intact GSL-microdomain feasible for further structural and functional characterization.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nagahori, N., Abe, M. & Nishimura, SI. Structural and functional glycosphingolipidomics by glycoblotting with aminooxy-functionalized gold nanoparticle. Nat Prec (2008). https://doi.org/10.1038/npre.2008.2034.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2008.2034.1