Abstract
Background:
Recognizing similarities and deriving relationships among protein molecules is a fundamentalrequirement in present-day biology. Similarities can be present at various levels which can be detected through comparison of protein sequences or their structural folds. In some cases similarities obscure at these levels could be present merely in the substructures at their binding sites. Inferring functional similarities between protein molecules by comparing their binding sites is still largely exploratory and not as yet a routine protocol. One ofthe main reasons for this is the limitation in the choice of appropriate analytical tools that can compare binding sites with high sensitivity. To benefit from the enormous amount of structural data that is being rapidly accumulated, it is essential to have high throughput tools that enable large scale binding site comparison.
Results:
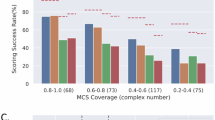
Here we present a new algorithm PocketMatch for comparison of binding sites in a frame invariantmanner. Each binding site is represented by 90 lists of sorted distances capturing shape and chemical nature of the site. The sorted arrays are then aligned using an incremental alignment method and scored to obtain PMScores for pairs of sites. A comprehensive sensitivity analysis and an extensive validation of the algorithm have been carried out. Perturbation studies where the geometry of a given site was retained but the residue types were changed randomly, indicated that chance similarities were virtually non-existent. Our analysis also demonstrates that shape information alone is insufficient to discriminate between diverse binding sites, unlesscombined with chemical nature of amino acids.
Conclusions:
A new algorithm has been developed to compare binding sites in accurate, efficient andhigh-throughput manner. Though the representation used is conceptually simplistic, we demonstrate that alongwith the new alignment strategy used, it is sufficient to enable binding comparison with high sensitivity. Novel methodology has also been presented for validating the algorithm for accuracy and sensitivity with respect to geometry and chemical nature of the site. The method is also fast and takes about 1/250th second for one comparison on a single processor. A parallel version on BlueGene has also been implemented.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kalidas, Y., Chandra, N. PocketMatch: A new algorithm to compare binding sites in protein structures. Nat Prec (2008). https://doi.org/10.1038/npre.2008.2142.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2008.2142.1