Abstract
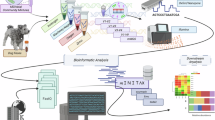
A major challenge for the development of resources for functional and comparative genomics is the extraction of data from the biomedical literature. Although text retrieval and extraction for biological data is an active research field, few applications have been integrated into production literature curation systems such as those of the model organism databases.In September 2008, Mouse Genome Informatics (MGI) at The Jackson Lab initiated a search for dictionary-based text mining tools that we could integrate into our curation workflow. MGI has rigorous document triage and annotation procedures designed to identify articles about mouse genome biology and determine whether those articles should be curated. We currently screens approximately 1000 journal articles a month for Gene Ontology terms, gene mapping, gene expression, phenotype data and other key biological information. Although we don’t foresee that human curation tasks can be fully automated in the near future, we are eager to implement entity name recognition and gene tagging tools that can help streamline our curation workflow and simplify gene indexing tasks in the MGI system. In this presentation, we discuss our search process and the steps we took to identify a short list of potential tools for further evaluation. We present our performance metrics and success criteria, and pilot projects in progress. The primary applications under current review are Fraunhofer SCAI’s ProMiner and NCBO’s Open-Biomedical Annotator.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dowell, K., McAndrews-Hill, M., Hill, D. et al. Integrating Text Mining into the MGI Biocuration Workflow. Nat Prec (2009). https://doi.org/10.1038/npre.2009.3262.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2009.3262.1