Abstract
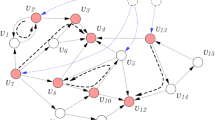
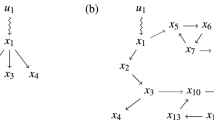
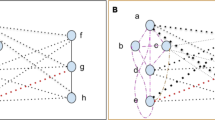
We present a novel algorithm that exhibits natural selection of paths in a network. If each node and weighted directed edge has a unique identifier, a path in the network is defined as an ordered list of these unique identifiers. We take a population perspective and view each path as a genotype. If each node has a node phenotype then a path phenotype is defined as the list of node phenotypes in order of traversal. We show that given appropriate path traversal, weight change and structural plasticity rules, a path is a unit of evolution because it can exhibit multiplicative growth (i.e. change it’s probability of being traversed), and have variation and heredity. Thus, a unit of evolution need not be a spatially distinct physical individual. The total set of paths in a network consists of all possible paths from the start node to a finish node. Each path phenotype is associated with a reward that determines whether the edges of that path will be multiplicatively strengthened (or weakened). A pair-wise tournament selection algorithm is implemented which compares the reward obtained by two paths. The directed edges of the winning path are strengthened, whilst the directed edges of the losing path are weakened. Edges shared by both paths are not changed (or weakened if diversity is desired). Each time a node is activated there is a probability that the path will mutate, i.e. find an alternative route that bypasses that node. This generates the potential for a novel but correlated path with a novel but correlated phenotype. By this process the more frequently traversed paths are responsible for most of the exploration. Nodes that are inactive for some period of time are lost (which is equivalent to connections to and from them being broken). This network-based natural selection compares favourably with a standard pair-wise tournament-selection based genetic algorithm on a range of combinatorial optimization problems and continuous parametric optimization problems. The network also exhibits memory of past selective environments and can store previously discovered characters for reuse in later optimization tasks. The pathway evolution algorithm has several possible implementations and permits natural selection with unlimited heredity without template replication.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fernando, C., Vasas, V., Szathmáry, E. et al. Natural Selection of Paths in Networks. Nat Prec (2011). https://doi.org/10.1038/npre.2011.5535.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2011.5535.1