Abstract
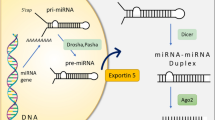
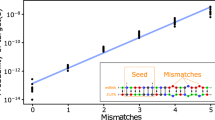
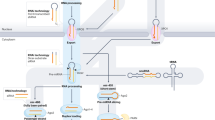
MicroRNAs (miRNAs) which are on average only 21-25 nucleotides long are key post-transcriptional regulators of gene expression in metazoans and plants. A proper quantitative understanding of miRNAs is required to comprehend their structures, functions, evolutions etc. In this paper, the nucleotide strings of miRNAs of three organisms namely Homo sapiens (hsa), Macaca mulatta (mml) and Pan troglodytes (ptr) have been quantified and classified based on some characterizing features. A network has been built up among the miRNAs for these three organisms through a class of discrete transformations namely Integral Value Transformations (IVTs), proposed by Sk. S. Hassan et al [1, 2]. Through this study we have been able to nullify or justify one given nucleotide string as a miRNA. This study will help us to recognize a given nucleotide string as a probable miRNA, without the requirement of any conventional biological experiment. This method can be amalgamated with the existing analysis pipelines, for small RNA sequencing data (designed for finding novel miRNA). This method would provide more confidence and would make the current analysis pipeline more efficient in predicting the probable candidates of miRNA for biological validation and filter out the improbable candidates.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hassan, S., Pal Choudhury, P., Goswami, A. et al. Quantification of miRNAs and Their Networks in the light of Integral Value Transformations. Nat Prec (2011). https://doi.org/10.1038/npre.2011.6035.2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2011.6035.2