Abstract
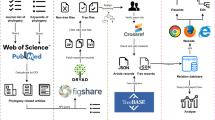
Sharing and re-use of data are essential to the progressive and self-correcting nature of science. In recognition of this principle, journals and funding agencies have adopted policies to encourage sharing of information ('data'), including empirical data as well as computed inferences such as phylogenetic trees.Here we summarize an ongoing analysis of 1) current practices for sharing phylogenetic trees and associated data; 2) current barriers to effective sharing and reuse of such data; and 3) prospects for reducing these barriers to promote more widespread sharing and re-use. Currently, the technical infrastructure is available to support (with some limitations) rudimentary archiving in conjunction with manuscript publication. Yet, most published trees are not archived, and there is no community standard governing the recommended format or content to ensure a re-usable phylogenetic record. Without a shift in emphasis toward re-usability, along with technology and standards to support such a shift, the value of trees (whether disseminated via public archives, or by other means) will be limited. Interviews with actual or potential secondary consumers of phylogenetic results suggest that there is a considerable market for re-use, but that most attempts end in disappointment. Phylogenetic results available via author requests, journal web sites, archival repositories and project web sites rarely include the critical information that secondary consumers seek, such as unique identifiers for biological sources (including species sources and accession numbers), indicators of quality, and documentation of the analytical methods used to obtain the results.Based on the analysis presented here, we suggest that enabling effective re-use entails a commitment by the research community to several changes from current practice: 1) using globally unique identifiers (GUIDs) to reference informational and material entities; 2) developing and using technology for documenting and exchanging the metadata that facilitate re-use; and 3) supporting development and use of a minimal reporting standard that indicates what data and metadata are considered essential for a re-useable phylogenetic record. We suggest that re-use may be catalyzed most rapidly by identifying and targeting (with appropriate technology) the most promising circumstances for re-use. These might include the extraction of sub-trees from large trees (for use in reconciliation, classification, and comparative analysis); the re-use of seed alignments, sub-alignments and homologized characters; the linking of phylogenies to geographic information (for use in ecology, phylogeography and biogeography); and the construction of supertrees and supermatrices.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
O’Meara, B., Whitacre, J., Mounce, R. et al. Publishing re-usable phylogenetic trees, in theory and practice. Nat Prec (2011). https://doi.org/10.1038/npre.2011.6048.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2011.6048.1