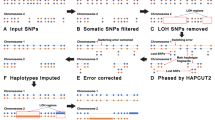
Abstract
Interpretation of allelic copy measurements at polymorphic markers in cancer samples presents distinctive challenges and opportunities. Due to frequent gross chromosomal alterations occurring in cancer (aneuploidy), many genomic regions are present at homologous-allele imbalance. Within such regions, the unequal contribution of alleles at heterozygous markers allows for direct phasing of the haplotype derived from each individual parent. In addition, genome-wide estimates of homologue specific copy- ratios (HSCRs) are important for interpretation of the cancer genome in terms of fixed integral copy-numbers. We describe HAPSEG, a probabilistic method to interpret bi- allelic marker data in cancer samples. HAPSEG operates by partitioning the genome into segments of distinct copy number and modeling the four distinct genotypes in each segment. We describe general methods for fitting these models to data which are suit- able for both SNP microarrays and massively parallel sequencing data. In addition, we demonstrate a specially tailored error-model for interpretation of systematic variations arising in microarray platforms. The ability to directly determine haplotypes from cancer samples represents an opportunity to expand reference panels of phased chromosomes, which may have general interest in various population genetic applications. In addition, this property may be exploited to interrogate the relationship between germline risk and cancer phenotype with greater sensitivity than is possible using unphased genotype. Finally, we exploit the statistical dependency of phased genotypes to enable the fitting of more elaborate sample-level error-model parameters, allowing more accurate estimation of HSCRs in cancer samples.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Carter, S., Meyerson, M. & Getz, G. Accurate estimation of homologue-specific DNA concentration-ratios in cancer samples allows long-range haplotyping. Nat Prec (2011). https://doi.org/10.1038/npre.2011.6494.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2011.6494.1
Keywords
This article is cited by
-
Chromosomal phase improves aneuploidy detection in non-invasive prenatal testing at low fetal DNA fractions
Scientific Reports (2022)
-
Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data
Nature Communications (2020)
-
Genomic characterization of genes encoding histone acetylation modulator proteins identifies therapeutic targets for cancer treatment
Nature Communications (2019)
-
Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition
Nature Communications (2016)
-
Deciphering clonality in aneuploid breast tumors using SNP array and sequencing data
Genome Biology (2014)