Abstract
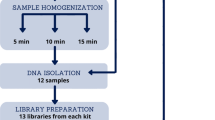
The objective of this publication is to provide the detailed protocol for metartancriptomisc studies of animal intestinal microbiota. The protocol describes isolation of high quality microbial community RNA from the mammalian intestinal content, subsequent mRNA enrichment, cDNA synthesis and sequencing. Twelve libraries were prepared, pooled in equimolar concentrations into a single library and sequenced on one GS Titanium 70 × 75 picotiter plate, following this protocol. The total number of reads obtained for 12 libraries was 1,155,062 (average 96,000 per library) and the combined size of 12 libraries was 521 million bases (average 43 million bases per library). The reported size of non-ribosomal RNA library fraction is ~15%, the fraction of non-ribosomal reads is ~17%. Hence we described a robust technique for metranscriptomic studies of animal intestinal microbiota. The double stranded cDNAs, prepared following this protocol, are suitable for pyrosequencing (454, Illumina), clone library construction or could be used to archive and store metaranscriptomic samples.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Poroyko, V., Hernandez, A., Alverdy, J. et al. Protocol for Metatranscriptomic analysis of Intestinal Microbiota. Nat Prec (2012). https://doi.org/10.1038/npre.2012.6934.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2012.6934.1