Abstract
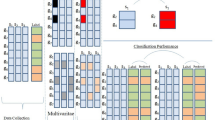
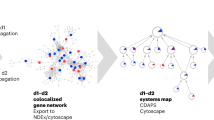
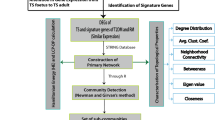
Simultaneous overexpression or underexpression of multiple genes, used in various forms as probes in the high-throughput microarray experiments, facilitates the identification of their underlying functional proximity. This kind of functional associativity (or conversely the separability) between the genes can be represented proficiently using co-expression networks. The extensive repository of diversified microarray data encounters a recent problem of multi-experimental data integration for the aforesaid purpose. This paper highlights a novel integration method of gene co-expression networks, based on the search for their consensus network, derived from diverse microarray experimental data for the purpose of clustering. The proposed methodology avoids the bias arising from missing value estimation. The method has been applied on microarray datasets arising from different category of experiments to integrate them. The consensus network, thus produced, reflects robustness based on biological validation.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bhattacharyya, M., Bandyopadhyay, S. Integration of Co-expression Networks for Gene Clustering. Nat Prec (2012). https://doi.org/10.1038/npre.2012.7126.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2012.7126.1