Key Points
-
The deubiquitylating enzyme (DUB) family contains ~100 proteins that remove the post-translational modification ubiquitin from a variety of substrates.
-
DUBs have key roles in various areas of cell biology of high relevance to pathologies such as autoimmune disorders, chronic inflammation, oncology and neurodegeneration.
-
DUBs are attractive targets for small-molecule drug discovery, as they contain a well-defined active site, and the majority of them have a catalytic cysteine.
-
Oxidative hydrolysis of the active-site cysteine is a challenge for DUB inhibitor screening, as reducing agents are often required to maintain DUB activity but frequently result in high false-positive rates if used at high concentrations.
-
Many of the reported DUB inhibitors have been shown to be rather non-selective in biochemical selectivity profiling assays.
-
Recent advances in screening substrates and technologies, as well as activity-based probes for monitoring target engagement, have facilitated progress in DUB drug discovery.
-
Increased understanding of DUB biology and emerging examples of potent and selective DUB inhibitors suggest that clinical development of DUB inhibitors is on the horizon.
Abstract
More than a decade after a Nobel Prize was awarded for the discovery of the ubiquitin–proteasome system and clinical approval of proteasome and ubiquitin E3 ligase inhibitors, first-generation deubiquitylating enzyme (DUB) inhibitors are now approaching clinical trials. However, although our knowledge of the physiological and pathophysiological roles of DUBs has evolved tremendously, the clinical development of selective DUB inhibitors has been challenging. In this Review, we discuss these issues and highlight recent advances in our understanding of DUB enzymology and biology as well as technological improvements that have contributed to the current interest in DUBs as therapeutic targets in diseases ranging from oncology to neurodegeneration.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ciechanover, A. The ubiquitin proteolytic system and pathogenesis of human diseases: a novel platform for mechanism-based drug targeting. Biochem. Soc. Trans. 31, 474–481 (2003).
Ciechanover, A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6, 79–87 (2005).
Gallastegui, N. & Groll, M. The 26S proteasome: assembly and function of a destructive machine. Trends Biochem. Sci. 35, 634–642 (2010).
Finley, D., Chen, X. & Walters, K. J. Gates, channels, and switches: elements of the proteasome machine. Trends Biochem. Sci. 41, 77–93 (2016).
Peth, A., Besche, H. C. & Goldberg, A. L. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol. Cell 36, 794–804 (2009).
Muratani, M. & Tansey, W. P. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4, 192–201 (2003).
Jesenberger, V. & Jentsch, S. Deadly encounter: ubiquitin meets apoptosis. Nat. Rev. Mol. Cell Biol. 3, 112–121 (2002).
Hicke, L. Protein regulation by monoubiquitin. Nat. Reviews Molecular Cell Biology 2, 195–201 (2001).
Jackson, S. P. & Durocher, D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 (2013). This review highlights how ubiquitylation and related processes control many cellular responses to DNA damage.
Husnjak, K. & Dikic, I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 (2012).
Herhaus, L. & Dikic, I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 16, 1071–1083 (2015).
Komander, D., Clague, M. J. & Urbe, S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 (2009).
van der Veen, A. G. & Ploegh, H. L. Ubiquitin-like proteins. Annu. Rev. Biochem. 81, 323–357 (2012).
Huang, C. J., Wu, D., Khan, A. F. . & Huo, L. J. DeSUMOylation: an important therapeutic target and protein regulatory event. DNA Cell Biol. 34, 652–660 (2015).
Murali, R., Wiesner, T. & Scolyer, R. A. Tumours associated with BAP1 mutations. Pathology 45, 116–126 (2013).
Oliveira, A. M. & Chou, M. M. USP6-induced neoplasms: the biologic spectrum of aneurysmal bone cyst and nodular fasciitis. Hum. Pathol. 45, 1–11 (2014).
Hao, Y. H. et al. USP7 acts as a molecular rheostat to promote WASH-dependent endosomal protein recycling and is mutated in a human neurodevelopmental disorder. Mol. Cell 59, 956–969 (2015).
Ma, Z. Y. et al. Recurrent gain-of-function USP8 mutations in Cushing's disease. Cell Res. 25, 306–317 (2015).
Reincke, M. et al. Mutations in the deubiquitinase gene USP8 cause Cushing's disease. Nat. Genet. 47, 31–38 (2015). References 18 and 19 are key publications identifying mutations in USP8 that enhance EGFR signalling and cause Cushing disease.
Homan, C. C. et al. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am. J. Hum. Genet. 94, 470–478 (2014).
Murtaza, M., Jolly, L. A., Gecz, J. & Wood, S. A. La FAM fatale: USP9X in development and disease. Cell. Mol. Life Sci. 72, 2075–2089 (2015).
Eichhorn, P. J. et al. USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma. Nat. Med. 18, 429–435 (2012).
Bignell, G. R. et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 25, 160–165 (2000).
Kawaguchi, Y. et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 8, 221–228 (1994).
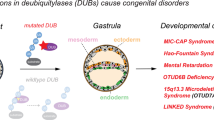
McDonell, L. M. et al. Mutations in STAMBP, encoding a deubiquitinating enzyme, cause microcephaly-capillary malformation syndrome. Nat. Genet. 45, 556–562 (2013).
Cohen, P. & Tcherpakov, M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell 143, 686–693 (2010). This is a key perspective highlighting DUBs as attractive drug targets.
Huang, X. & Dixit, V. M. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 26, 484–498 (2016). This recent review highlights drug discovery efforts targeting the ubiquitin system, including DUBs.
Sacco, J. J., Coulson, J. M., Clague, M. J. & Urbe, S. Emerging roles of deubiquitinases in cancer-associated pathways. IUBMB Life 62, 140–157 (2010).
Wang, L. & Dent, S. Y. Functions of SAGA in develo-pment and disease. Epigenomics 6, 329–339 (2014).
Nicholson, B. & Suresh Kumar, K. G. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem. Biophys. 60, 61–68 (2011).
Cremona, C. A., Sancho, R., Diefenbacher, M. E. & Behrens, A. Fbw7 and its counteracting forces in stem cells and cancer: Oncoproteins in the balance. Semin. Cancer Biol. 36, 52–61 (2016).
D'Arcy, P., Wang, X. & Linder, S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol. Ther. 147, 32–54 (2015).
Souroullas, G. P. & Sharpless, N. E. Stem cells: Down's syndrome link to ageing. Nature 501, 325–326 (2013).
van Loosdregt, J. et al. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity 39, 259–271 (2013). First report demonstrating that USP7 deubiquitylates and stabilizes FOXP3 in T reg cells.
Sun, H. et al. Bcr-Abl ubiquitination and Usp9x inhibition block kinase signaling and promote CML cell apoptosis. Blood 117, 3151–3162 (2011).
Savio, M. G. et al. USP9X controls EGFR fate by deubiquitinating the endocytic adaptor Eps15. Curr. Biol. 26, 173–183 (2016).
Richardson, P. G., Hideshima, T. & Anderson, K. C. Bortezomib (PS-341): a novel, first-in-class proteasome inhibitor for the treatment of multiple myeloma and other cancers. Cancer Control 10, 361–369 (2003).
Chen, D., Frezza, M., Schmitt, S., Kanwar, J. & Dou, Q. P. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr. Cancer Drug Targets 11, 239–253 (2011).
Yao, T. & Cohen, R. E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 (2002).
Song, Y. et al. Deubiquitylating enzyme Rpn11/POH1/PSMD14 As therapeutic target in multiple myeloma. Blood 128, 4469–4469 (2016).
Wang, B. et al. POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat. Commun. 6, 8704 (2015).
Liu, H., Buus, R., Clague, M. J. & Urbe, S. Regulation of ErbB2 receptor status by the proteasomal DUB POH1. PLoS ONE 4, e5544 (2009).
Kakarougkas, A. et al. Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection. Nucleic Acids Res. 41, 10298–10311 (2013).
Hu, M. et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J 24, 3747–3756 (2005).
Eletr, Z. M. & Wilkinson, K. D. Regulation of proteolysis by human deubiquitinating enzymes. Biochim. Biophys. Acta 1843, 114–128 (2014).
Lee, B. H. et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 (2010).
Wu, N. et al. Over-expression of deubiquitinating enzyme USP14 in lung adenocarcinoma promotes proliferation through the accumulation of beta-catenin. Int. J. Mol. Sci. 14, 10749–10760 (2013).
Wang, Y. et al. Ubiquitin-specific protease 14 (USP14) regulates cellular proliferation and apoptosis in epithelial ovarian cancer. Med. Oncol. 32, 379 (2015).
Xu, D. et al. Phosphorylation and activation of ubiquitin-specific protease-14 by Akt regulates the ubiquitin-proteasome system. eLife 4, e10510 (2015).
Jung, H. et al. Deubiquitination of Dishevelled by Usp14 is required for Wnt signaling. Oncogenesis 2, e64 (2013).
Qiu, X. B. et al. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 25, 5742–5753 (2006).
Yao, T. et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 8, 994–1002 (2006).
Koulich, E., Li, X. & DeMartino, G. N. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol. Biol. Cell 19, 1072–1082 (2008).
Lam, Y. A., Xu, W., DeMartino, G. N. & Cohen, R. E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385, 737–740 (1997).
Mazumdar, T. et al. Regulation of NF-kappaB activity and inducible nitric oxide synthase by regulatory particle non-ATPase subunit 13 (Rpn13). Proc. Natl Acad. Sci. USA 107, 13854–13859 (2010).
Wang, L. et al. High expression of UCH37 is significantly associated with poor prognosis in human epithelial ovarian cancer. Tumour Biol. 35, 11427–11433 (2014).
Fang, Y. et al. Ubiquitin C-terminal hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating PRP19. Biochim. Biophys. Acta 1833, 559–572 (2013).
Richardson, P. G. A review of the proteasome inhibitor bortezomib in multiple myeloma. Expert Opin. Pharmacother. 5, 1321–1331 (2004).
Wang, X. et al. Synthesis and evaluation of derivatives of the proteasome deubiquitinase inhibitor b-AP15. Chem. Biol. Drug Design 86, 1036–1048 (2015).
D'Arcy, P. et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 17, 1636–1640 (2011).
Berndtsson, M. et al. Induction of the lysosomal apoptosis pathway by inhibitors of the ubiquitin-proteasome system. Int. J. Cancer 124, 1463–1469 (2009).
Tian, Z. et al. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood 123, 706–716 (2014).
Zhou, H.-J. et al. Compositions and methods for JAMM protein inhibition. WO2012158435 (A1) (2012).
Zhou, H.-J., Parlati, F. & Wustrow, D. Methods and compositions for JAMM protease inhibition. WO2013123071 (A1) (2013).
Zhou, H.-J. & Wustrow, D. Compositions and methods for JAMM protein inhibition. WO2014066506 (A2) (2014).
O'Connor, M. J. Targeting the DNA Damage response in cancer. Mol. Cell 60, 547–560 (2015).
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Jacq, X., Kemp, M., Martin, N. M. & Jackson, S. P. Deubiquitylating enzymes and DNA damage response pathways. Cell Biochem. Biophys. 67, 25–43 (2013).
Castella, M. et al. FANCI Regulates recruitment of the FA core complex at sites of DNA damage independently of FANCD2. PLoS Genet. 11, e1005563 (2015).
Nijman, S. M. et al. The Deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 17, 331–339 (2005).
Huang, T. T. et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8, 339–347 (2006).
Guervilly, J. H., Renaud, E., Takata, M. & Rosselli, F. USP1 deubiquitinase maintains phosphorylated CHK1 by limiting its DDB1-dependent degradation. Hum. Mol. Genet. 20, 2171–2181 (2011).
Williams, S. A. et al. USP1 Deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 146, 918–930 (2011).
Cohn, M. A. et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell 28, 786–797 (2007).
Chen, J. et al. Selective and cell-active inhibitors of the USP1/ UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem. Biol. 18, 1390–1400 (2011).
Ritorto, M. S. et al. Screening of DUB activity and specificity by MALDI-TOF mass spectrometry. Nat. Commun. 5, 4763 (2014). Key publication assessing substrate specificity of 42 DUBs and selectivity profiling of 9 reported DUB inhibitors against a panel of 32 DUBs.
Dexheimer, T. S. et al. Synthesis and structure-activity relationship studies of N-benzyl-2-phenylpyrimidin-4-amine derivatives as potent USP1/UAF1 deubiquitinase inhibitors with anticancer activity against nonsmall cell lung cancer. J. Med. Chem. 57, 8099–8110 (2014).
Liang, Q. et al. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 10, 298–304 (2014). Identification of the USP1 inhibitor ML323 and demonstration that the compound potentiates cisplatin cytotoxicity in non-small-cell lung cancer and osteosarcoma cells.
Schoenfeld, A. R., Apgar, S., Dolios, G., Wang, R. & Aaronson, S. A. BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol. Cell. Biol. 24, 7444–7455 (2004).
Wiltshire, T. D. et al. Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J. Biol. Chem. 285, 14565–14571 (2010).
Orthwein, A. et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426 (2015).
Burkhart, R. A. et al. Mitoxantrone targets human ubiquitin-specific peptidase 11 (USP11) and is a potent inhibitor of pancreatic cancer cell survival. Mol. Cancer Res. 11, 901–911 (2013).
Wijnhoven, P. et al. USP4 Auto-Deubiquitylation Promotes Homologous Recombination. Mol. Cell 60, 362–373 (2015).
McGarry, E. et al. The deubiquitinase USP9X maintains DNA replication fork stability and DNA damage checkpoint responses by regulating CLASPIN during S-phase. Cancer Res. 76, 2384–2393 (2016).
Wolfsperger, F. et al. Deubiquitylating enzyme USP9x regulates radiosensitivity in glioblastoma cells by Mcl-1-dependent and -independent mechanisms. Cell Death Dis. 7, e2039 (2016).
Kapuria, V. et al. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 70, 9265–9276 (2010).
Kapuria, V. et al. A novel small molecule deubiquitinase inhibitor blocks Jak2 signaling through Jak2 ubiquitination. Cell Signal 23, 2076–2085 (2011).
Clague, M. J. et al. Deubiquitylases from genes to organism. Physiol. Rev. 93, 1289–1315 (2013).
Prives, C. Signaling to p53: breaking the MDM2 p53 circuit. Cell 95, 5–8 (1998).
Harris, S. L. & Levine, A. J. The p53 pathway: positive and negative feedback loops. Oncogene 24, 2899–2908 (2005).
Li, M. et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416, 648–653 (2002).
Li, M., Brooks, C. L., Kon, N. & Gu, W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 13, 879–886 (2004).
Cummins, J. M. & Vogelstein, B. HAUSP is required for p53 destabilization. Cell Cycle 3, 689–692 (2004).
van der Horst, A. et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 8, 1064–1073 (2006).
Song, M. S. et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455, 813–817 (2008).
Lecona, E. et al. USP7 is a SUMO deubiquitinase essential for DNA replication. Nat. Struct. Mol. Biol. 23, 270–277 (2016).
Colland, F. et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 8, 2286–2295 (2009).
Reverdy, C. et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem. Biol. 19, 467–477 (2012).
Chauhan, D. et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell 22, 345–358 (2012).
Fan, Y. H. et al. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 4, e867 (2013).
Gavory, G. et al. Discovery and characterization of novel, highly potent and selective USP7 inhibitors. Cancer Res. 75 (Suppl. 15), abstr. LB-257 (2015).
Liu, H. et al. The Machado–Joseph disease deubiquitinase ataxin-3 regulates the stability and apoptotic function of p53. PLoS Biol. 14, e2000733 (2016).
Cuella-Martin, R. et al. 53BP1 integrates DNA repair and p53-dependent cell fate decisions via distinct mechanisms. Mol. Cell 64, 51–64 (2016).
Popov, N. et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 9, 765–774 (2007).
Diefenbacher, M. E. et al. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J. Clin. Invest. 124, 3407–3418 (2014).
Biju, M. et al. Discovery of potent and selective small molecule USP20 inhibitors. Presented at Ubiquitin Drug Discovery and Diagnostics Conference, Philadelphia, 2012.
Mizuno, E. et al. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell 16, 5163–5174 (2005).
Niendorf, S. et al. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol. Cell. Biol. 27, 5029–5039 (2007).
Yewale, C., Baradia, D., Vhora, I., Patil, S. & Misra, A. Epidermal growth factor receptor targeting in cancer: a review of trends and strategies. Biomaterials 34, 8690–8707 (2013).
Colombo, M. et al. Synthesis and biological evaluation of 9-oxo-9H-indeno[1,2-b]pyrazine-2,3-dicarbonitrile analogues as potential inhibitors of deubiquitinating enzymes. ChemMedChem 5, 552–558 (2010).
Byun, S. et al. USP8 is a novel target for overcoming gefitinib resistance in lung cancer. Clin. Cancer Res. 19, 3894–3904 (2013).
Inui, M. et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat. Cell Biol. 13, 1368–1375 (2011).
Wilkinson, K. D. et al. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science 246, 670–673 (1989).
Day, I. N. & Thompson, R. J. UCHL1 (PGP 9.5): neuronal biomarker and ubiquitin system protein. Progress Neurobiol. 90, 327–362 (2010).
Hurst-Kennedy, J., Chin, L. S. & Li, L. Ubiquitin C-Terminal Hydrolase L1 in Tumorigenesis. Biochem. Res. Int. 2012, 123706 (2012).
Hussain, S. et al. The de-ubiquitinase UCH-L1 is an oncogene that drives the development of lymphoma in vivo by deregulating PHLPP1 and Akt signaling. Leukemia 24, 1641–1655 (2010).
Kim, H. J. et al. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene 28, 117–127 (2009).
Jang, M. J., Baek, S. H. & Kim, J. H. UCH-L1 promotes cancer metastasis in prostate cancer cells through EMT induction. Cancer Lett. 302, 128–135 (2011).
Gu, Y. Y. et al. The de-ubiquitinase UCHL1 promotes gastric cancer metastasis via the Akt and Erk1/2 pathways. Tumour Biol. 36, 8379–8387 (2015).
Liu, Y. et al. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem. Biol. 10, 837–846 (2003).
Mermerian, A. H., Case, A., Stein, R. L. & Cuny, G. D. Structure-activity relationship, kinetic mechanism, and selectivity for a new class of ubiquitin C-terminal hydrolase-L1 (UCH-L1) inhibitors. Bioorg. Med. Chem. Lett. 17, 3729–3732 (2007).
Davies, C. W. et al. The co-crystal structure of ubiquitin carboxy-terminal hydrolase L1 (UCHL1) with a tripeptide fluoromethyl ketone (Z-VAE(OMe)-FMK). Bioorg. Med. Chem. Lett. 22, 3900–3904 (2012).
Jones, A. et al. Novel Compounds. WO2016046530 (2016). First patent from Mission Therapeutics describing its DUB inhibitors.
Kemp, M., Stockley, M. & Jones, A. Cyanopyrrolidines as DUB inhibitors for the treatment of cancers. WO2017009650 (2017).
Hussain, S., Bedekovics, T., Chesi, M., Bergsagel, P. L. & Galardy, P. J. UCHL1 is a biomarker of aggressive multiple myeloma required for disease progression. Oncotarget 6, 40704–40718 (2015).
Schrecengost, R. S. et al. USP22 regulates oncogenic signaling pathways to drive lethal cancer progression. Cancer Res. 74, 272–286 (2014).
Liu, Y. L., Yang, Y. M., Xu, H. & Dong, X. S. Aberrant expression of USP22 is associated with liver metastasis and poor prognosis of colorectal cancer. J. Surg. Oncol. 103, 283–289 (2011).
Zhang, Y. et al. Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. J. Cancer Res. Clin. Oncol. 137, 1245–1253 (2011).
Li, J., Wang, Z. & Li, Y. USP22 nuclear expression is significantly associated with progression and unfavorable clinical outcome in human esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 138, 1291–1297 (2012).
Piao, S. et al. USP22 is useful as a novel molecular marker for predicting disease progression and patient prognosis of oral squamous cell carcinoma. PLoS ONE 7, e42540 (2012).
Zhang, J. et al. Identification of the E3 deubiquitinase ubiquitin-specific peptidase 21 (USP21) as a positive regulator of the transcription factor GATA3. J. Biol. Chem. 288, 9373–9382 (2013). This publication demonstrates that USP21 deubiquitylates and stabilizes the transcription factor GATA3 in T reg cells.
Li, Y. et al. USP21 prevents the generation of T-helper-1-like Treg cells. Nat. Commun. 7, 13559 (2016).
Facciabene, A., Motz, G. T. & Coukos, G. T-Regulatory cells: key players in tumor immune escape and angiogenesis. Cancer Res. 72, 2162–2171 (2012).
Ross, C. A. & Pickart, C. M. The ubiquitin-proteasome pathway in Parkinson's disease and other neurodegenerative diseases. Trends Cell Biol. 14, 703–711 (2004).
Todi, S. V. & Paulson, H. L. Balancing act: deubiquitinating enzymes in the nervous system. Trends Neurosci. 34, 370–382 (2011).
Ristic, G., Tsou, W. L. & Todi, S. V. An optimal ubiquitin-proteasome pathway in the nervous system: the role of deubiquitinating enzymes. Frontiers Mol. Neurosci. 7, 72 (2014).
Green, D. R., Galluzzi, L. & Kroemer, G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science 333, 1109–1112 (2011).
Ross, J. M., Olson, L. & Coppotelli, G. Mitochondrial and ubiquitin proteasome system dysfunction in ageing and disease: two sides of the same coin? Int. J. Mol. Sci. 16, 19458–19476 (2015).
Martin, I., Dawson, V. L. & Dawson, T. M. Recent advances in the genetics of Parkinson's disease. Annu. Rev. Genom. Hum. Genet. 12, 301–325 (2011).
Hauser, D. N. & Hastings, T. G. Mitochondrial dysfunction and oxidative stress in Parkinson's disease and monogenic parkinsonism. Neurobiol. Dis. 51, 35–42 (2013).
Narendra, D. P. & Youle, R. J. Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid. Redox Signal. 14, 1929–1938 (2011).
Bingol, B. et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510, 370–375 (2014). This key publication demonstrates that USP30 antagonizes mitophagy driven by the ubiquitin ligase parkin and protein kinase PINK1.
Durcan, T. M. & Fon, E. A. The three 'P's of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev. 29, 989–999 (2015).
Nakamura, N. & Hirose, S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Mol. Biol. Cell 19, 1903–1911 (2008).
Narendra, D., Tanaka, A., Suen, D. F. & Youle, R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 (2008).
Yue, W. et al. A small natural molecule promotes mitochondrial fusion through inhibition of the deubiquitinase USP30. Cell Res. 24, 482–496 (2014).
Jones, A., Kemp, M., Stockley, M., Gibson, K. R. & Whitlock, G. A. Cyano-pyrrolidine compounds as USP30 inhibitors. WO2016156816 (2016).
Durcan, T. M. et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 33, 2473–2491 (2014).
Alexopoulou, Z. et al. Deubiquitinase Usp8 regulates alpha-synuclein clearance and modifies its toxicity in Lewy body disease. Proc. Natl Acad. Sci. USA 113, E4688–E4697 (2016).
Cornelissen, T. et al. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum. Mol. Genet. 23, 5227–5242 (2014).
Durcan, T. M., Kontogiannea, M., Bedard, N., Wing, S. S. & Fon, E. A. Ataxin-3 deubiquitination is coupled to Parkin ubiquitination via E2 ubiquitin-conjugating enzyme. J. Biol. Chem. 287, 531–541 (2012).
Guterman, A. & Glickman, M. H. Deubiquitinating enzymes are IN/(trinsic to proteasome function). Curr. Protein Peptide Sci. 5, 201–211 (2004).
Finley, D., Gahman, T. C., King, R. W., Lee, B. H. & Lee, M. J. Tricyclic proteasome activity enhancing compounds. WO 2012012712 (2012).
Chambers, R. J., Foley, M. & Tait, B. Proteasome activity modulating compounds. WO 2013112651 (2013).
Chambers, R. J., Foley, M. & Tait, B. Proteasome activity enhancing compounds. WO2013112699 (2013). References 154 and 155 are Proteostasis Therapeutics patents that describe USP14 inhibitors.
Crimmins, S. et al. Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. J. Neurosci. 26, 11423–11431 (2006).
Ortuno, D., Carlisle, H. J. & Miller, S. Does inactivation of USP14 enhance degradation of proteasomal substrates that are associated with neurodegenerative diseases? F1000Res. 5, 137 (2016).
Joo, H. Y. et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature 449, 1068–1072 (2007).
Adorno, M. et al. Usp16 contributes to somatic stem-cell defects in Down's syndrome. Nature 501, 380–384 (2013). This work demonstrates that USP16 has an important role in antagonizing the self-renewal and/or senescence pathways in Down syndrome.
Xu, J. C., Dawson, V. L. & Dawson, T. M. Usp16: key controller of stem cells in Down syndrome. EMBO J. 32, 2788–2789 (2013).
Hu, H. & Sun, S. C. Ubiquitin signaling in immune responses. Cell Res. 26, 457–483 (2016). A recent review that describes how ubiquitylation regulates immune signalling and discusses of roles of many DUBs.
Jiang, X. & Chen, Z. J. The role of ubiquitylation in immune defence and pathogen evasion. Nat. Rev. Immunology 12, 35–48 (2011).
Adhikari, A., Xu, M. & Chen, Z. J. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26, 3214–3226 (2007).
Tokunaga, F. Linear ubiquitination-mediated NF-kappaB regulation and its related disorders. J. Biochem. 154, 313–323 (2013).
Harhaj, E. W. & Dixit, V. M. Deubiquitinases in the regulation of NF-kappaB signaling. Cell Res. 21, 22–39 (2011).
Catrysse, L., Vereecke, L., Beyaert, R. & van Loo, G. A20 in inflammation and autoimmunity. Trends Immunol. 35, 22–31 (2014).
Wertz, I. E. et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430, 694–699 (2004). This publication demonstrates that phosphorylation of A20 promotes cleavage of K63-linked polyubiquitin chains.
Skaug, B. et al. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol. Cell 44, 559–571 (2011).
Wertz, I. E. et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature 528, 370–375 (2015).
Zhang, M. et al. Regulation of IkappaB kinase-related kinases and antiviral responses by tumor suppressor CYLD. J. Biol. Chem. 283, 18621–18626 (2008).
Friedman, C. S. et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 9, 930–936 (2008).
Fiil, B. K. & Gyrd-Hansen, M. Met1-linked ubiquitination in immune signalling. FEBS J. 281, 4337–4350 (2014).
Iwai, K., Fujita, H. & Sasaki, Y. Linear ubiquitin chains: NF-kappaB signalling, cell death and beyond. Nat. Rev. Mol. Cell Biol. 15, 503–508 (2014).
MacDuff, D. A. et al. Phenotypic complementation of genetic immunodeficiency by chronic herpesvirus infection. eLife https://dx.doi.org/10.7554/eLife.04494.002 (2015).
Gerlach, B. et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 (2011).
Ikeda, F. et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature 471, 637–641 (2011).
Tokunaga, F. et al. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636 (2011).
Tokunaga, F. et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat. Cell Biol. 11, 123–132 (2009).
Boisson, B. et al. Human HOIP and LUBAC deficiency underlies autoinflammation, immunodeficiency, amylopectinosis, and lymphangiectasia. J. Exp. Med. 212, 939–951 (2015).
Boisson, B. et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nature Immunol. 13, 1178–1186 (2012).
Keusekotten, K. et al. OTULIN Antagonizes LUBAC Signaling by Specifically Hydrolyzing Met1-Linked Polyubiquitin. Cell 153, 1312–1326 (2013).
Rivkin, E. et al. The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature 498, 318–324 (2013). Together with reference 181, this is a key publication that identifies OTULIN as the DUB that removes linear ubiquitin chains.
Damgaard, R. B. et al. The Deubiquitinase OTULIN is an essential negative regulator of inflammation and autoimmunity. Cell 166, 1215–1230.e1220 (2016).
Honke, N., Shaabani, N., Zhang, D. E., Hardt, C. & Lang, K. S. Multiple functions of USP18. Cell Death Dis. 7, e2444 (2016).
Rioux, J. D. & Abbas, A. K. Paths to understanding the genetic basis of autoimmune disease. Nature 435, 584–589 (2005).
Walsh, K. P. & Mills, K. H. Dendritic cells and other innate determinants of T helper cell polarisation. Trends Immunol. 34, 521–530 (2013).
Bettelli, E., Oukka, M. & Kuchroo, V. K. TH-17 cells in the circle of immunity and autoimmunity. Nature Immunol. 8, 345–350 (2007).
Bettelli, E., Korn, T., Oukka, M. & Kuchroo, V. K. Induction and effector functions of TH17 cells. Nature 453, 1051–1057 (2008).
Yang, J. et al. Cutting edge: Ubiquitin-specific protease 4 promotes Th17 cell function under inflammation by deubiquitinating and stabilizing RORgammat. J. Immunol. 194, 4094–4097 (2015).
Okada, K. et al. Vialinin A is a ubiquitin-specific peptidase inhibitor. Bioorg. Med. Chem. Lett. 23, 4328–4331 (2013).
Jin, J. et al. Epigenetic regulation of the expression of Il12 and Il23 and autoimmune inflammation by the deubiquitinase Trabid. Nature Immunol. 17, 259–268 (2016).
Liu, X. et al. USP18 inhibits NF-kappaB and NFAT activation during Th17 differentiation by deubiquitinating the TAK1-TAB1 complex. J. Exp. Med. 210, 1575–1590 (2013).
Hu, H. et al. Otud7b facilitates T cell activation and inflammatory responses by regulating Zap70 ubiquitination. J. Exp. Med. 213, 399–414 (2016).
Wang, H. et al. ZAP-70: an essential kinase in T-cell signaling. Cold Spring Harb. Perspect. Biol. 2, a002279 (2010).
Damsker, J. M., Hansen, A. M. & Caspi, R. R. Th1 and Th17 cells: adversaries and collaborators. Ann. NY Acad. Sci. 1183, 211–221 (2010).
Pan, L. et al. Deubiquitination and stabilization of T-bet by USP10. Biochem. Biophys. Res. Commun. 449, 289–294 (2014).
Bailey-Elkin, B. A., van Kasteren, P. B., Snijder, E. J., Kikkert, M. & Mark, B. L. Viral OTU deubiquitinases: a structural and functional comparison. PLoS Pathog. 10, e1003894 (2014).
Sun, L. et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE 7, e30802 (2012).
Ratia, K. et al. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl Acad. Sci. USA 105, 16119–16124 (2008).
Bailey-Elkin, B. A. et al. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 289, 34667–34682 (2014).
Lei, J. et al. Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features. Antiviral Res. 109, 72–82 (2014).
Frias-Staheli, N. et al. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2, 404–416 (2007).
Pruneda, J. N. et al. The molecular basis for ubiquitin and ubiquitin-like specificities in bacterial effector proteases. Mol. Cell 63, 261–276 (2016).
Artavanis-Tsakonas, K. et al. Identification by functional proteomics of a deubiquitinating/deNeddylating enzyme in Plasmodium falciparum. Mol. Microbiol. 61, 1187–1195 (2006).
Frickel, E. M. et al. Apicomplexan UCHL3 retains dual specificity for ubiquitin and Nedd8 throughout evolution. Cell. Microbiol. 9, 1601–1610 (2007).
Artavanis-Tsakonas, K. et al. Characterization and structural studies of the Plasmodium falciparum ubiquitin and Nedd8 hydrolase UCHL3. J. Biol. Chem. 285, 6857–6866 (2010).
Wrigley, J. D. et al. Enzymatic characterisation of USP7 deubiquitinating activity and inhibition. Cell Biochem. Biophys. 60, 99–111 (2011).
Sahtoe, D. D. & Sixma, T. K. Layers of DUB regulation. Trends Biochem. Sci. 40, 456–467 (2015). This review discusses the extent and variety of allosteric regulation modes of DUB catalytic activity.
Mevissen, T. E. T. & Komander, D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192 (2017). A very recent and comprehensive review that describes DUB substrate specificity and regulation of catalytic activity.
Kemp, M. Recent advances in the discovery of deubiquitinating enzyme inhibitors. Progress Med. Chem. 55, 149–192 (2016).
Kathman, S. G., Xu, Z. & Statsyuk, A. V. A fragment-based method to discover irreversible covalent inhibitors of cysteine proteases. J. Med. Chem. 57, 4969–4974 (2014).
Donato, N. J. et al. Deubiquitinase inhibitors and methods for use of the same. WO2015054555 (2015).
Turk, B. Targeting proteases: successes, failures and future prospects. Nat. Rev. Drug Discov. 5, 785–799 (2006).
Hershko, A., Ciechanover, A., Heller, H., Haas, A. L. & Rose, I. A. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl Acad. Sci. USA 77, 1783–1786 (1980).
Nijman, S. M. et al. A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 (2005).
Faesen, A. C. et al. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol. Cell 44, 147–159 (2011).
Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009).
Meyer, H. & Weihl, C. C. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J. Cell Sci. 127, 3877–3883 (2014).
Kato, J. Y. & Yoneda-Kato, N. Mammalian COP9 signalosome. Genes Cells 14, 1209–1225 (2009).
Sahtoe, D. D., van Dijk, W. J., Ekkebus, R., Ovaa, H. & Sixma, T. K. BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat. Commun. 7, 10292 (2016).
Ventii, K. H. & Wilkinson, K. D. Protein partners of deubiquitinating enzymes. Biochem. J. 414, 161–175 (2008).
D'Andrea, A. Inhibitors of USP1 deubiquitinating enzyme complex. US2008167229 (2008).
Guedat, P. et al. Novel tetracyclic inhibitors of cysteine proteases, the pharmaceutical compositions thereof and their therapeutic applications. US2008103149 (2008).
Colland, F. & Gourdel, M. E. Selective and reversible inhibitors of ubiquitin specific protease 7. WO2013030218 (2013).
Dang, L. C., Melandri, F. D. & Stein, R. L. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry 37, 1868–1879 (1998).
de Jong, A. et al. Ubiquitin-based probes prepared by total synthesis to profile the activity of deubiquitinating enzymes. Chembiochem 13, 2251–2258 (2012).
McGouran, J. F. et al. Fluorescence-based active site probes for profiling deubiquitinating enzymes. Org. Biomol. Chem. 10, 3379–3383 (2012).
Orcutt, S. J., Wu, J., Eddins, M. J., Leach, C. A. & Strickler, J. E. Bioluminescence assay platform for selective and sensitive detection of Ub/Ubl proteases. Biochim. Biophys. Acta 1823, 2079–2086 (2012).
Leach, C. A., Strickler, J. E. & Eddins, M. J. Methods of screening for inhibitors of enzymes. WO2013043970 (2013).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012). A landmark review of the broad scope of ubiquitin modifications, illustrating the large diversity of substrates handled by DUBs.
Hospenthal, M. K., Mevissen, T. E. & Komander, D. Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest). Nat. Protoc. 10, 349–361 (2015).
Sowa, M. E., Bennett, E. J., Gygi, S. P. & Harper, J. W. Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009). This work describes the extensive landscape of protein–protein interactions engaged by DUBs.
Harrigan, J. & Jacq, X. Monitoring Target Engagement of Deubiquitylating Enzymes Using Activity Probes: Past, Present, and Future. Methods Mol. Biol. 1449, 395–410 (2016). A recent review of the utility and benefits of activity probes to monitor DUB target engagement in cells and tissues.
Hewings, D. S., Flygare, J. A., Bogyo, M. & Wertz, I. E. Activity-based probes for the ubiquitin conjugation-deconjugation machinery: new chemistries, new tools, and new insights. FEBS J. 284, 1555–1576 (2017).
Borodovsky, A. et al. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 20, 5187–5196 (2001).
Galardy, P., Ploegh, H. L. & Ovaa, H. Mechanism-based proteomics tools based on ubiquitin and ubiquitin-like proteins: crystallography, activity profiling, and protease identification. Methods Enzymol. 399, 120–131 (2005).
El Oualid, F. et al. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew. Chem. Int. Ed. 49, 10149–10153 (2010).
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011).
Altun, M. et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 18, 1401–1412 (2011). Detailed analysis of DUB activity using ubiquitin-based activity probes and characterization of the selectivity of the DUB inhibitors PR-619 and P2077.
Wang, X. et al. The 19S Deubiquitinase inhibitor b-AP15 is enriched in cells and elicits rapid commitment to cell death. Mol. Pharmacol. 85, 932–945 (2014).
Chen, Y. et al. Expression and clinical significance of UCH37 in human esophageal squamous cell carcinoma. Dig. Dis. Sci. 57, 2310–2317 (2012).
Gray, D. A. et al. Elevated expression of Unph, a proto-oncogene at 3p21.3, in human lung tumors. Oncogene 10, 2179–2183 (1995).
Zhang, L. et al. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-beta type I receptor. Nat. Cell Biol. 14, 717–726 (2012).
Heo, M. J. et al. microRNA-148a dysregulation discriminates poor prognosis of hepatocellular carcinoma in association with USP4 overexpression. Oncotarget 5, 2792–2806 (2014).
Bayraktar, S. et al. USP-11 as a predictive and prognostic factor following neoadjuvant therapy in women with breast cancer. Cancer J. 19, 10–17 (2013).
Huether, R. et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat. Commun. 5, 3630 (2014).
Ma, M. & Yu, N. Ubiquitin-specific protease 7 expression is a prognostic factor in epithelial ovarian cancer and correlates with lymph node metastasis. Onco Targets Ther. 9, 1559–1569 (2016).
Masuya, D. et al. The HAUSP gene plays an important role in non-small cell lung carcinogenesis through p53-dependent pathways. J. Pathol. 208, 724–732 (2006).
Zhao, G. Y. et al. USP7 overexpression predicts a poor prognosis in lung squamous cell carcinoma and large cell carcinoma. Tumour Biol. 36, 1721–1729 (2015).
Forbes, S. A. et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D777–D783 (2017).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Zheng, S. et al. Heterogeneous expression and biological function of ubiquitin carboxy-terminal hydrolase-L1 in osteosarcoma. Cancer Lett. 359, 36–46 (2015).
Akishima-Fukasawa, Y. et al. Significance of PGP9.5 expression in cancer-associated fibroblasts for prognosis of colorectal carcinoma. Am. J. Clin. Pathol. 134, 71–79 (2010).
Ma, Y. et al. Proteomic profiling of proteins associated with lymph node metastasis in colorectal cancer. J. Cell. Biochem. 110, 1512–1519 (2010).
Goto, Y. et al. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1alpha. Nat. Commun. 6, 6153 (2015).
Gunia, S. et al. Protein gene product 9.5 is diagnostically helpful in delineating high-grade renal cell cancer involving the renal medullary/sinus region from invasive urothelial cell carcinoma of the renal pelvis. Hum. Pathol. 44, 712–717 (2013).
Otsuki, T. et al. Expression of protein gene product 9.5 (PGP9.5)/ubiquitin-C-terminal hydrolase 1 (UCHL-1) in human myeloma cells. Br. J. Haematol. 127, 292–298 (2004).
Hibi, K. et al. PGP9.5 as a candidate tumor marker for non-small-cell lung cancer. Am. J. Pathol. 155, 711–715 (1999).
Hu, J. et al. Expression patterns of USP22 and potential targets BMI-1, PTEN, p-AKT in non-small-cell lung cancer. Lung cancer 77, 593–599 (2012).
Liang, J. X. et al. Ubiquitin specific protease 22-induced autophagy is correlated with poor prognosis of pancreatic cancer. Oncol. Rep. 32, 2726–2734 (2014).
Cunningham, C. N. et al. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat. Cell Biol. 17, 160–169 (2015).
Wilson, S. M. et al. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat. Genet. 32, 420–425 (2002).
Ceriani, M., Amigoni, L., D'Aloia, A., Berruti, G. & Martegani, E. The deubiquitinating enzyme UBPy/USP8 interacts with TrkA and inhibits neuronal differentiation in PC12 cells. Exp. Cell Res. 333, 49–59 (2015).
Daviet, L. & Colland, F. Targeting ubiquitin specific proteases for drug discovery. Biochimie 90, 270–283 (2008).
Bruzzone, F., Vallarino, M., Berruti, G. & Angelini, C. Expression of the deubiquitinating enzyme mUBPy in the mouse brain. Brain Res. 1195, 56–66 (2008).
Yang, W. et al. The histone H2A deubiquitinase Usp16 regulates embryonic stem cell gene expression and lineage commitment. Nat. Commun. 5, 3818 (2014).
Ovaa, H. et al. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc. Natl Acad. Sci. USA 101, 2253–2258 (2004).
Liu, L. Q. et al. A novel ubiquitin-specific protease, UBP43, cloned from leukemia fusion protein AML1-ETO-expressing mice, functions in hematopoietic cell differentiation. Mol. Cell. Biol. 19, 3029–3038 (1999).
Malakhova, O. A. et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 25, 2358–2367 (2006).
Zhong, B. et al. Ubiquitin-specific protease 25 regulates TLR4-dependent innate immune responses through deubiquitination of the adaptor protein TRAF3. Sci. Signal 6, ra35 (2013).
Lin, D. et al. Induction of USP25 by viral infection promotes innate antiviral responses by mediating the stabilization of TRAF3 and TRAF6. Proc. Natl Acad. Sci. USA 112, 11324–11329 (2015).
Ren, Y. et al. The Type I Interferon-IRF7 Axis Mediates Transcriptional Expression of Usp25 Gene. J. Biol. Chem. 291, 13206–13215 (2016).
Liu, Y. C., Penninger, J. & Karin, M. Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunology 5, 941–952 (2005).
Wertz, I. & Dixit, V. A20—a bipartite ubiquitin editing enzyme with immunoregulatory potential. Adv. Exp. Med. Biol. 809, 1–12 (2014).
Wang, L. et al. USP4 positively regulates RIG-I-mediated antiviral response through deubiquitination and stabilization of RIG-I. J. Virol. 87, 4507–4515 (2013).
Fan, Y. H. et al. USP4 targets TAK1 to downregulate TNFalpha-induced NF-kappaB activation. Cell Death Differ. 18, 1547–1560 (2011).
Han, L. et al. The E3 deubiquitinase USP17 is a positive regulator of retinoic acid-related orphan nuclear receptor gammat (RORgammat) in Th17 cells. J. Biol. Chem. 289, 25546–25555 (2014).
Ni, Y. et al. The deubiquitinase USP17 regulates the stability and nuclear function of IL-33. Int. J. Mol. Sci. 16, 27956–27966 (2015).
Lim, K. H., Ramakrishna, S. & Baek, K. H. Molecular mechanisms and functions of cytokine-inducible deubiquitinating enzymes. Cytokine Growth Factor Rev. 24, 427–431 (2013).
Annunziata, C. M. et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12, 115–130 (2007).
Keats, J. J. et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell 12, 131–144 (2007).
Hellerbrand, C. et al. Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis 28, 21–27 (2007).
Massoumi, R. et al. Down-regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J. Exp. Med. 206, 221–232 (2009).
Ye, Y. et al. TRE17/USP6 oncogene translocated in aneurysmal bone cyst induces matrix metalloproteinase production via activation of NF-kappaB. Oncogene 29, 3619–3629 (2010).
Alwan, H. A. & van Leeuwen, J. E. UBPY-mediated epidermal growth factor receptor (EGFR) de-ubiquitination promotes EGFR degradation. J. Biol. Chem. 282, 1658–1669 (2007).
Harbour, J. W. et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330, 1410–1413 (2010).
O'Shea, S. J. et al. A population-based analysis of germline BAP1 mutations in melanoma. Hum. Mol. Genet. 26, 717–728 (2017).
Testa, J. R. et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat. Genet. 43, 1022–1025 (2011).
Popova, T. et al. Germline BAP1 mutations predispose to renal cell carcinomas. Am. J. Hum. Genet. 92, 974–980 (2013).
Li, Z., Wang, D., Messing, E. M. & Wu, G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 6, 373–378 (2005).
Malakhov, M. P., Malakhova, O. A., Kim, K. I., Ritchie, K. J. & Zhang, D. E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277, 9976–9981 (2002).
Zhong, H. et al. Ubiquitin-specific proteases 25 negatively regulates virus-induced type I interferon signaling. PLoS ONE 8, e80976 (2013).
Colleran, A. et al. Deubiquitination of NF-kappaB by ubiquitin-specific protease-7 promotes transcription. Proc. Natl Acad. Sci. USA 110, 618–623 (2013).
Zhou, F. et al. Ubiquitin-specific protease 4 mitigates Toll-like/interleukin-1 receptor signaling and regulates innate immune activation. J. Biol. Chem. 287, 11002–11010 (2012).
Chen, R. et al. The ubiquitin-specific protease 17 is involved in virus-triggered type I IFN signaling. Cell Res. 20, 802–811 (2010).
Wang, X. et al. The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells. Scientif. Rep. 6, 26979 (2016).
Maloney, D. J. et al. Inhibitors of the USP1/UAF1 deubiquitinase complex and uses thereof. WO2014105952 (A2) (2014).
Weinstock, J. et al. Selective dual inhibitors of the cancer-related deubiquitylating proteases USP7 and USP47. Acs Med. Chem. Lett. 3, 789–792 (2012).
Foley, M., Tait, B. & Cullen, M. Proteostasis regulators. WO2012154967 (A1) (2012).
Finley, D., King, R. W., Lee, B. H., Lee, M. J. & Gahman, T. C. Compositions and methods for enhancing proteasome activity. WO2011094545 (A2) (2011).
Davis, M. I. et al. Small Molecule inhibition of the ubiquitin-specific protease USP2 accelerates cyclin D1 degradation and leads to cell cycle arrest in colorectal cancer and mantle cell lymphoma models. J. Biol. Chem. 291, 24628–24640 (2016).
Liu, J. et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147, 223–234 (2011).
Zhong, B. et al. Negative regulation of IL-17-mediated signaling and inflammation by the ubiquitin-specific protease USP25. Nature Immunol. 13, 1110–1117 (2012).
Acknowledgements
The authors thank K. Dry for extensive editing and expert advice, and other members of the S.P.J. academic laboratory for helpful discussions and advice. The authors thank L. Greger for the generation of the phylogenic tree. Research in the S.P.J. laboratory is funded by the Cancer Research UK (CRUK) Program Grant C6/A18796 and a Wellcome Trust Investigator Award (206388/Z/17/Z), and institute core infrastructure funding is provided by the CRUK (C6946/A24843) and the Wellcome Trust (WT203144).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
S.P.J. is part-time employed by Mission Therapeutics, Cambridge, UK, owns shares and is a director of the company. J.A.H. is a full-time employee of Mission Therapeutics. N.M.M. and X.J. own shares in Mission Therapeutics.
Related links
Glossary
- Ubiquitin
-
A small protein that is conjugated to other proteins (including itself) as a post-translational modification, often to control cellular signalling or degradation of the modified protein.
- Ubiquitin-like proteins
-
(UBLs). Proteins such as small ubiquitin-like modifier (SUMO) or interferon-stimulated gene 15 (ISG15) that adopt a β-grasp fold, which is characteristic of ubiquitin and related proteins.
- Proteasome
-
A protein complex that recognizes and degrades polyubiquitylated proteins. The 19S regulatory subunit contains ATPase- and ubiquitin-binding sites, and the 20S catalytic core contains proteases.
- Deubiquitylating enzymes
-
(DUBs). Proteins that remove ubiquitin from substrate proteins or cleave ubiquitin precursors.
- Tumour suppressor
-
A type of protein that normally restricts the proliferation or invasive properties of normal cells. Mutations or loss-of-expression of tumour suppressors can lead to cancer progression.
- Oncogene
-
A type of protein that normally controls growth, differentiation or survival of cells but whose mutation or expression at abnormally high levels promotes tumorigenesis.
- Isopeptidase
-
A type of enzyme that hydrolyses amide bonds that occur outside of the main chain in a polypeptide chain.
- Regulatory T cells
-
(Treg cells). A subpopulation of T cells that modulate the immune system, maintain tolerance to self-antigens and prevent autoimmune disease.
- Mitochondria
-
Cellular organelles that produce ATP through oxidative phosphorylation.
- Mitophagy
-
The process of eliminating damaged and aged mitochondria via sequestration and hydrolytic degradation by the lysosome.
- ax J
-
A hypomorphic allele of USP14. The ataxia mutation (axJ) is a recessive neurological mutation that results in reduced growth, ataxia and hindlimb muscle wasting in mice.
- Cytokines
-
Members of a class of immunoregulatory proteins (interleukins or interferons) that are secreted by cells, especially cells of the immune system.
- T helper cells
-
(TH cells). Activated T cells linked to defence against various types of pathogens, which are commonly defined by their production of particular cytokines. The best-characterized effector T cell subsets include TH1, TH2 and TH17 cells.
- Allosteric
-
Binding to a site other than the active site of the enzyme, resulting in modulation of enzymatic activity.
- Activity-based probes
-
(ABPs). Tools that incorporate elements for targeting, modification and/or detection of labelled proteins, enabling assessment of enzymatic activity and inhibition.
Rights and permissions
About this article
Cite this article
Harrigan, J., Jacq, X., Martin, N. et al. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov 17, 57–78 (2018). https://doi.org/10.1038/nrd.2017.152
Published:
Issue date:
DOI: https://doi.org/10.1038/nrd.2017.152
This article is cited by
-
The emerging role of E3 ubiquitin ligases and deubiquitinases in metabolic dysfunction-associated steatotic liver disease
Journal of Translational Medicine (2025)
-
NLRP3 inflammasome-modulated angiogenic function of EPC via PI3K/ Akt/mTOR pathway in diabetic myocardial infarction
Cardiovascular Diabetology (2025)
-
Role of USP7 in the regulation of tolerogenic dendritic cell function in type 1 diabetes
Cellular & Molecular Biology Letters (2025)
-
Epigenetic regulation and post-translational modifications of ferroptosis-related factors in cardiovascular diseases
Clinical Epigenetics (2025)
-
Post-translational modifications in anaplastic thyroid carcinoma: biological mechanisms and therapeutic potential
Cancer Cell International (2025)