Key Points
-
Genome-wide association studies (GWAS) have been highly successful in the analyses of human genomic data. The increased availability of microorganism whole genomes provides the opportunity for microbial GWAS.
-
Initial microbial GWAS have had success identifying variants for traits under strong selection, such as drug resistance, in a range of bacteria, viruses and protozoa.
-
Several challenges to microbial GWAS exist that could hinder identifying variants under moderate selection. The primary challenge is the increased population stratification in microorganisms owing to selection and complex recombination patterns.
-
Novel software that is tailored to the needs of microbial GWAS would greatly expedite progress in the field. In particular, the application of polygenic methods has yet to be evaluated in microorganisms.
-
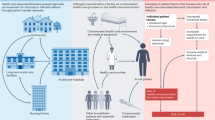
An exciting future area of research is the generation of host and microbial genomics data within the same samples. This will allow for genome-to-genome analyses to test for host–microorganism interactions.
Abstract
The reduced costs of sequencing have led to whole-genome sequences for a large number of microorganisms, enabling the application of microbial genome-wide association studies (GWAS). Given the successes of human GWAS in understanding disease aetiology and identifying potential drug targets, microbial GWAS are likely to further advance our understanding of infectious diseases. These advances include insights into pressing global health problems, such as antibiotic resistance and disease transmission. In this Review, we outline the methodologies of GWAS, the current state of the field of microbial GWAS, and how lessons from human GWAS can direct the future of the field.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Wood, A. R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Bush, W. S. & Moore, J. H. Chapter 11: Genome-wide association studies. PLoS Comput. Biol. 8, e1002822 (2012). This review discusses in detail the methods, nuances and caveats of GWAS.
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Manolio, T. A. Bringing genome-wide association findings into clinical use. Nat. Rev. Genet. 14, 549–558 (2013).
Reich, D. E. & Lander, E. S. On the allelic spectrum of human disease. Trends Genet. 17, 502–510 (2001).
Hirschhorn, J. N. & Daly, M. J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108 (2005).
Cordell, H. J. Detecting gene–gene interactions that underlie human diseases. Nat. Rev. Genet. 10, 392–404 (2009).
Thomas, D. Gene–environment-wide association studies: emerging approaches. Nat. Rev. Genet. 11, 259–272 (2010).
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010).
Wang, K. et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 17, 1665–1674 (2007).
Bansal, V., Libiger, O., Torkamani, A. & Schork, N. J. Statistical analysis strategies for association studies involving rare variants. Nat. Rev. Genet. 11, 773–785 (2010).
Lees, J. A. et al. Sequence element enrichment analysis to determine the genetic basis of bacterial phenotypes. Nat. Commun. 7, 12797 (2016). This methods paper presents a mixed model approach to microbial GWAS, including the analysis of k-mers.
Earle, S. G. et al. Identifying lineage effects when controlling for population structure improves power in bacterial association studies. Nat. Microbiol. 1, 16041 (2016). This methods paper presents an approach to disentangling the effects of single SNPs and lineage effects within microbial GWAS.
Ioannidis, J. P., Thomas, G. & Daly, M. J. Validating, augmenting and refining genome-wide association signals. Nat. Rev. Genet. 10, 318–329 (2009).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Novembre, J. et al. Genes mirror geography within Europe. Nature 456, 98–101 (2008).
Didelot, X. & Maiden, M. C. Impact of recombination on bacterial evolution. Trends Microbiol. 18, 315–322 (2010).
Read, T. D. & Massey, R. C. Characterizing the genetic basis of bacterial phenotypes using genome-wide association studies: a new direction for bacteriology. Genome Med. 6, 109 (2014). The authors present an important review of the findings of bacterial GWAS.
Rosenberg, N. A. et al. Genome-wide association studies in diverse populations. Nat. Rev. Genet. 11, 356–366 (2010).
Farhat, M. R. et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet. 45, 1183–1189 (2013). This microbial GWAS introduces the PhyC method, which uses phylogenetic trees to carry out a genome-wide scan of convergent evolution.
Dudbridge, F. & Gusnanto, A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 32, 227–234 (2008).
NCI–NHGRI Working Group on Replication in Association Studies et al. Replicating genotype-phenotype associations. Nature 447, 655–660 (2007).
Zollner, S. & Pritchard, J. K. Overcoming the winner's curse: estimating penetrance parameters from case-control data. Am. J. Hum. Genet. 80, 605–615 (2007).
Manolio, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Zeggini, E. & Ioannidis, J. P. Meta-analysis in genome-wide association studies. Pharmacogenomics 10, 191–201 (2009).
Visscher, P. M., Brown, M. A., McCarthy, M. I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012). An important perspective on the lessons learnt from human GWAS and predictions of the future of the field.
Wray, N. R. et al. Pitfalls of predicting complex traits from SNPs. Nat. Rev. Genet. 14, 507–515 (2013). A useful review of a range of polygenic methods and their applications.
Purcell, S. M. et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009).
Lee, S. H., Yang, J., Goddard, M. E., Visscher, P. M. & Wray, N. R. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics 28, 2540–2542 (2012).
Visscher, P. M. & Yang, J. A plethora of pleiotropy across complex traits. Nat. Genet. 48, 707–708 (2016).
Tan, J. C. et al. An optimized microarray platform for assaying genomic variation in Plasmodium falciparum field populations. Genome Biol. 12, R35 (2011).
Cheeseman, I. H. et al. A major genome region underlying artemisinin resistance in malaria. Science 336, 79–82 (2012).
Alam, M. T. et al. Dissecting vancomycin-intermediate resistance in Staphylococcus aureus using genome-wide association. Genome Biol. Evol. 6, 1174–1185 (2014).
Chewapreecha, C. et al. Comprehensive identification of single nucleotide polymorphisms associated with β-lactam resistance within pneumococcal mosaic genes. PLoS Genet. 10, e1004547 (2014).
Malaria Genomic Epidemiology Network. A global network for investigating the genomic epidemiology of malaria. Nature 456, 732–737 (2008).
Pillay, D. et al. PANGEA-HIV: phylogenetics for generalised epidemics in Africa. Lancet Infect. Dis. 15, 259–261 (2015).
Desjardins, C. A. et al. Genomic and functional analyses of Mycobacterium tuberculosis strains implicate ald in D-cycloserine resistance. Nat. Genet. 48, 544–551 (2016).
Miotto, O. et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 47, 226–234 (2015).
Sheppard, S. K. et al. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc. Natl Acad. Sci. USA 110, 11923–11927 (2013).
Bartha, I. et al. A genome-to-genome analysis of associations between human genetic variation, HIV-1 sequence diversity, and viral control. eLife 2, e01123 (2013). An example of a genome-to-genome analysis with both host and microbial GWAS data.
Laabei, M. et al. Predicting the virulence of MRSA from its genome sequence. Genome Res. 24, 839–849 (2014).
Power, R. A. et al. Genome-wide association study of HIV whole genome sequences validated using drug resistance. PLoS ONE 11, e0163476 (2016).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Chen, P. E. & Shapiro, B. J. The advent of genome-wide association studies for bacteria. Curr. Opin. Microbiol. 25, 17–24 (2015).
Thornton, T. & McPeek, M. S. ROADTRIPS: case-control association testing with partially or completely unknown population and pedigree structure. Am. J. Hum. Genet. 86, 172–184 (2010).
Lippert, C. et al. FaST linear mixed models for genome-wide association studies. Nat. Methods 8, 833–837 (2011).
Evangelou, E. & Ioannidis, J. P. Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet. 14, 379–389 (2013).
McCarthy, M. I. et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9, 356–369 (2008).
Traylor, M. et al. Using phenotypic heterogeneity to increase the power of genome-wide association studies: application to age at onset of ischaemic stroke subphenotypes. Genet. Epidemiol. 37, 495–503 (2013).
Power, R. A. et al. Genome-wide association for major depression through age at onset stratification: major depressive disorder working group of the Psychiatric Genomics Consortium. Biol. Psychiatry http://dx.doi.org/10.1016/j.biopsych.2016.05.010 (2016).
Hamshere, M. L. et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol. Psychiatry 18, 708–712 (2013).
Rietveld, C. A. et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340, 1467–1471 (2013).
Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379 (2013).
Chapman, S. J. & Hill, A. V. Human genetic susceptibility to infectious disease. Nat. Rev. Genet. 13, 175–188 (2012).
Bartha, I. et al. Estimating the respective contributions of human and viral genetic variation to HIV control. Preprint at bioRxivhttp://dx.doi.org/10.1101/029017 (2015).
Walker, T. M. et al. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect. Dis. 15, 1193–1202 (2015).
Fraser, C. et al. Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science 343, 1243727 (2014).
Acknowledgements
Research supported by a South African MRC Flagship grant (MRC-RFA-UFSP-01-2013/UKZN HIVEPI), Wellcome Trust grants (098051 and 201355/Z/16/Z) and a Royal Society Newton Advanced Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Genome-wide association studies
-
(GWAS). A hypothesis-free method that tests hundreds of thousands of variants across the genome to identify alleles that are associated with a phenotype.
- Single-nucleotide polymorphisms
-
(SNPs). A base position where two alleles exist with a frequency of >1% in the population.
- Heritability
-
The proportion of phenotypic variance that is due to inherited genetic variation.
- Beta
-
The standardized regression coefficient, derived from linear regressions in genome-wide association studies of continuous traits. It is reported as an estimate of the effect size of a single-nucleotide polymorphism (SNP), and reflects the change in phenotype expected from carrying a copy of the reference allele of the SNP.
- Odds ratio
-
(OR). The typical means of reporting the effect size of a single-nucleotide polymorphism in a case–control (or other binary phenotype) genome-wide association study. It is derived from a logistic regression, and represents the odds of the phenotype when carrying the reference allele, compared with the odds of the phenotype in the absence of the reference allele.
- Main effects
-
The effects of a variant on the phenotype without accounting for any possible interactions with other variants or environmental factors.
- Epistatic interactions
-
Interactions between variants at different locations in the genome.
- Power
-
The probability that an analysis will reject the null hypothesis when the alternative hypothesis is true. It is influenced by numerous factors, such as the effect size and sample size.
- Linkage disequilibrium
-
(LD). Correlations between variants due to co-inheritance. LD is usually higher between variants that are closer together, and is broken down by recombination.
- Phred scores
-
A measure of the quality of sequencing at a given locus, specifically the confidence in the calling of alleles at that locus.
- K-mers
-
A sequence of bases of length k that, in microbial genome-wide association studies, can be used as the genetic variant tested for association with the phenotype.
- Superinfection
-
When an individual is infected with multiple strains of the same microorganism.
- False positives
-
Variants, or any other predictors, that are identified as significantly associated with a phenotype but that are not causal. In the case of genome-wide association studies, this is usually due to confounding from population structure or insufficient quality control.
- Clonal
-
The case in which reproduction produces genetically identical organisms, and so does not introduce novel variants or recombination.
- Panmictic
-
A population in which clonal structure has been lost due to frequent recombination.
- Genome-wide significance
-
The P value cut-off for declaring a variant significantly associated with a phenotype, accounting for the number of variants tested and the correlations between them.
- Effect size
-
The proportion of variance in a phenotype predicted by a variant.
- Polygenic methods
-
Statistical approaches that focus on the combined effects of many genetic variants rather than on the effect of any individual variant.
- Pleiotropic
-
Pleiotropic variants are those that have an effect on multiple distinct phenotypes.
Rights and permissions
About this article
Cite this article
Power, R., Parkhill, J. & de Oliveira, T. Microbial genome-wide association studies: lessons from human GWAS. Nat Rev Genet 18, 41–50 (2017). https://doi.org/10.1038/nrg.2016.132
Published:
Issue date:
DOI: https://doi.org/10.1038/nrg.2016.132
This article is cited by
-
Genomic variants distinguish Pseudomonas aeruginosa from cystic fibrosis sputum and bloodstream infection
BMC Infectious Diseases (2026)
-
Genome co-adaptation and the evolution of methicillin resistant Staphylococcus aureus
Genome Biology (2026)
-
Partially connected neural networks for complex trait prediction: application to human height
Human Genetics (2026)
-
aurora: a machine learning gwas tool for analyzing microbial habitat adaptation
Genome Biology (2025)
-
Identification of genetic determinants of antibiotic resistance in Helicobacter pylori isolates in Vietnam by high-throughput sequencing
BMC Microbiology (2025)