Abstract
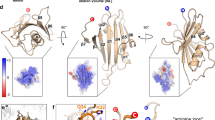
Many viruses regulate translation of polycistronic mRNA using a -1 ribosomal frameshift induced by an RNA pseudoknot. A pseudoknot has two stems that form a quasi-continuous helix and two connecting loops. A 1.6 Å crystal structure of the beet western yellow virus (BWYV) pseudoknot reveals rotation and a bend at the junction of the two stems. A loop base is inserted in the major groove of one stem with quadruple-base interactions. The second loop forms a new minor-groove triplex motif with the other stem, involving 2'-OH and triple-base interactions, as well as sodium ion coordination. Overall, the number of hydrogen bonds stabilizing the tertiary interactions exceeds the number involved in Watson–Crick base pairs. This structure will aid mechanistic analyses of ribosomal frameshifting.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Gesteland, R.F. & Atkins, J.F. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 65 , 741–768 (1996).
Farabaugh, P.J. Programmed translational frameshifting. Microbiol. Rev. 60, 103–134 (1996).
Jacks, T., Madhani, H.D., Masiarz, F.R. & Varmus, H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell 55, 447–458 (1988).
Chamorro, M., Parkin, N. & Varmus, H.E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc. Natl. Acad. Sci. USA 89, 713–717 (1992).
ten Dam, E., Brierley, I., Inglis, S. & Pleij, C. Identification and analysis of the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus-1. Nucleic Acids Res. 22 , 2304–2310 (1994).
Brierley, I., Rolley, N.J., Jenner, A.J. & Inglis, S.C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 220, 889–902 (1991).
Tzeng T.-H., Tu, C.-L. & Bruenn J.A. Ribosomal frameshifting requires a pseudoknot in the Saccharomyces cerevisiae double-stranded RNA virus. J. Virol. 66, 999–1006 ( 1992).
Miller, W.A., Dinesh-Kumar, S.P. & Paul, C.P. Luteovirus gene expression. Crit. Rev. Plant Sci. 14, 179–211 ( 1995).
Somogyi, P., Jenner, A.J., Brierley, I. & Inglis, S.C. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 13, 6931–6940 (1993).
Pleij, C.W.A., Rietveld, K. & Bosch, L. A new principle of RNA folding on pseudoknotting. Nucleic Acids Res. 13, 1717–1731 (1985).
Chen, X. et al. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 14, 842–852 ( 1995).
ten Dam, E.B., Verlaan, P.W.G. & Pleij, C.W.A. Analysis of the role of the pseudoknot component in the SRV-1 gag-pro ribosomal frameshift signal: loop lengths and stability of the stem regions. RNA 1, 146 –154 (1995).
Puglisi, J.D., Wyatt, J.R. & Tinoco, I. Jr. Conformation of an RNA pseudoknot. J. Mol. Biol. 214, 437– 453 (1990).
Shen, L.X. & Tinoco, I. Jr. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J. Mol. Biol. 247, 963– 978 (1995).
Du, Z., Giedroc, D.P. & Hoffman, D.W. Structure of the autoregulatory pseudoknot within the gene 32 messenger RNA of bacteriophage T2 and T6: a model for a possible family of structurally related pseudoknots. Biochemistry 35, 4187–4198 (1996).
Du, Z., Holland, J.A., Hansen, M.R., Giedroc, D.P. & Hoffman, D.W. Basepairings within the RNA pseudoknot associated with simian retrovirus-1 gag-pro frameshift site. J. Mol. Biol. 270, 464–470 (1997).
Kang, H. & Tinoco, I. Jr. A mutant RNA pseudoknot that promotes ribosomal frameshifting in mouse mammary tumor virus. Nucleic Acids Res. 25, 1943–1949 (1997).
Kolk, M.H. et al. NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA. Science 280, 434 –438 (1998).
Saenger, W. Principles of nucleic acid structure. (Springer-Verlag, New York; 1984).
Felsenfeld, G., Davies, D.R. & Rich, A. Formation of a three-stranded polynucleotide molecule. J. Amer. Chem. Soc. 79, 2023– 2024 (1957).
Rajagopal, P. & Feigon, J. Triple-strand formation in the homopurine : homopyrimidine DNA oligonucleotides d(G-A)4 and d(T-C)4 . Nature 339, 637– 640 (1989).
Egli, M. & Gessner, R.V. Stereoelectronic effects of deoxyribose O4' on DNA conformation. Proc. Natl. Acad. Sci. USA 92, 180–184 (1995).
Pley, H.W., Flaherty, K.M. & McKay, D.B. Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature 372, 111–113 ( 1994).
Cate, J. et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273, 1678– 1685 (1996).
Brown, T. & Hunter, W.N. Non-Watson–Crick base associations in DNA and RNA revealed by single crystal x-ray diffraction methods: mismatches, modified bases, and nonduplex DNA. Biopoly. 44, 91–103 (1997).
Pley, H.W., Flaherty, K.M. & McKay D.B. Three-dimensional structure of a hammerhead ribozyme. Nature 372, 68–74 (1994).
Scott, W.G., Finch, J.T. & Klug, A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for catalytic cleavage. Cell 81 , 991–1002 (1995).
Quigley, G.J. & Rich, A. Structural domains of transfer RNA molecules. Science 194, 796– 806 (1976).
Portmann, S., Grimm, S., Workman, C., Usman, N. & Egli, M. Crystal structures of an A-form duplex with single-adenosine bulges and conformational basis for site specific RNA self-cleavage. Chem. Biol. 3, 173–184 (1996).
Klimasauskas, S., Kumar, S., Roberts, R.J. & Cheng, X. Hhal methyltransferase flips its target base out of the DNA helix. Cell 76 , 357–369 (1994).
Hartman, K.A. & Rich, A. The tautomeric form of helical polyribocytidylic acids. J. Am. Chem. Soc. 87, 2033– 2039 (1965).
Gehring, K., Leroy, J.-L., Guéron, M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature 363, 561– 565 (1993).
Chen, L., Cai, L., Zhang, X. & Rich, A. Crystal structure of a four-stranded intercalated DNA: d(C4). Biochemistry 33, 13540–13546 (1994).
Wyatt, J.R., Puglisi, J.D. & Tinoco, I. Jr. RNA pseudoknots. Stability and loop size requirements. J. Mol. Biol. 214, 455 –470 (1990).
Gluick, T.C., Wills, N.M., Gesteland, R.F. & Draper, D.E. Folding of an mRNA pseudoknot required for stop codon readthrough: effects of mono- and divalent ions on stability. Biochemistry 36, 16173–16186 (1997).
Tu, C., Tzeng, T.-H. & Bruenn, J.A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl. Acad. Sci. USA 89, 8636–8640 (1992).
Sung, D. & Kang, H. Mutational analysis of the RNA pseudoknot involved in efficient ribosomal frameshifting in simian retrovirus-1. Nucleic Acids Res. 26, 1369–1372 (1998).
Jaeger, J., Restle T. & Steitz, T.A. The structure of HIV-1 reverse transcriptase complexed with an RNA pseudoknot inhibitor. EMBO J. 17, 4535–4542 (1998).
Pleij, C.W.A. RNA Pseudoknots. Curr. Opin. Struct. Biol. 4, 337–344 (1994).
Ortoleva-Donnelly, L., Szewczak, A.A., Gutell, R.R. & Strobel S.A. The chemical basis of adenosine conservation throughout the Tetrahymena ribozyme. RNA 4, 498– 519 (1998).
Dinman, J.D., Ruiz-Echevarria, M.J. & Peltz, S.W. Translating old drugs into new treatments: ribosomal frameshifting as a target for antiviral agents. Trends. Biotechnol. 16, 190–196 ( 1998).
Milligan, J.F. & Uhlenbeck, O.C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180, 51–62 (1989).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol . 276, 307–326 ( 1997).
CCP4. Collaborative computing project number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 ( 1994).
de La Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 ( 1997).
Jones, T.A., Zou, J.-Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building models in electron density maps and location of errors in these models. Acta Crystallogr. A 47, 110– 119 (1991).
Brünger, A.T. X-PLOR Version 3.1: a system for X-ray crystallography and NMR. (Yale University Press, New Haven; 1992).
Pann, N.S. & Read, R.J. Improved structure refinement through maximum likelihood. Acta Crystallogr. A 52, 659–668 (1996).
Lavery, R. & Sklenar, H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dynam. 6, 63–91 (1988).
Carson, M. Ribbons 2.0. J. Appl. Crystallogr. 24, 958 –961 (1991).
Acknowledgements
We thank E. de La Fortelle for extensive help on the program SHARP; G. Minasov and I. Berger for help in data processing and refinement, Y.-G. Kim for frameshifting discussions; C. Ogata (X4A-NSLS), M. Soltis (SSRL) and the staff at CHESS F2 for synchrotron assistance. We acknowledge help from T. Schwartz, D. Chan, M. Schade and K.-J. Wu on data collection; V. Tereshko, A. and H. Szoke for technical assistance; J. Cate, D. Bartel, M. Rould, U. RajBhandary, A. Herbert and B. Brown for helpful discussions. We are indebted to H. Taube and S. Holbrook for providing us with the osmium hexammine triflate compound; J. McCloskey and P. Crain for bromine mass spectroscopy analysis. We also thank L. Gerhke, K. Lowenhaupt and F. Houser-Scott for assistance in the T7 transcription system; D. Lauffenburger and S. Zhang at the MIT Center of Biomedical Engineering for providing the computer facility. This research was supported by grants from the National Science Foundation, the National Institutes of Health, the National Foundation for Cancer Research and the National Aeronautics and Space Administration.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Su, L., Chen, L., Egli, M. et al. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat Struct Mol Biol 6, 285–292 (1999). https://doi.org/10.1038/6722
Received:
Accepted:
Issue date:
DOI: https://doi.org/10.1038/6722
This article is cited by
-
Novel base triples in RNA structures revealed by graph theoretical searching methods
BMC Bioinformatics (2011)
-
A processed noncoding RNA regulates an altruistic bacterial antiviral system
Nature Structural & Molecular Biology (2011)
-
Structure of the SAM-II riboswitch bound to S-adenosylmethionine
Nature Structural & Molecular Biology (2008)
-
Viral RNA pseudoknots: versatile motifs in gene expression and replication
Nature Reviews Microbiology (2007)