Abstract
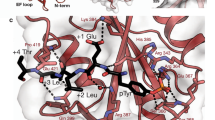
Crystal structures of the amino-terminal SH2 domain of the p85α subunit of phosphatidylinositol (PI) 3-kinase, alone and in complex with phosphopeptides bearing pTyr-Met/Val-Xaa-Met motifs, show that phosphopeptides bind in the two-pronged manner seen in high-affinity Lck and Src SH2 complexes, with conserved interactions between the domain and the peptide segment from phosphotyrosine to Met+3. Peptide binding requires the rearrangement of a tyrosyl side chain in the BG loop to create the hydrophobic Met+3 binding pocket. The structures suggest a mechanism for the biological specificity exhibited by PI 3-kinase in its interactions with phosphoprotein partners.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Kapeller, R. & Cantley, L.C. Phosphatidylinositol 3-kinase. Bioessays 16, 565–576 (1994).
Yao, R. & Cooper, G.M. Requirement for phosphatidyinositol-3 kinase in the prevention of apotosis by nerve growth factor. Science 267, 2003–2005 (1995).
Skolnik, E.Y. et al. Cloning of PI3 kinase-associated p85 utilizing a novel method for expression/cloning of target proteins for receptor tyrosine kinases. Cell 65, 83–90 (1991).
Otsu, M. et al. Characterization of two 85 kD proteins that associate with receptor tyrosine kinases, middle-T/pp60c-src complexes and PI3-kinase. Cell 65, 91–104 (1991).
Escobedo, J.A. et al. cDNA cloning of a novel 85 kD protein that has SH2 domains and regulates binding of PI3-kinase to the PDGF b-receptor. Cell 65, 75–82 (1991).
Klippel, A., Escobedo, J.A., Hu, Q. & Williams, L.T. A region of the 85-kda subunit of phosphatidylinositol 3-kinase binds the 110-kda catalytic subunit in vivo. Mol. Cell. Biol. 13, 5560–5566 (1994).
Dhand, R. et al. PI 3-kinase - structural and functional analysis of intersubunit interactions. Embo J. 13, 511–521 (1994).
Hu, P., Mondino, A., Skolnik, E.Y. & J., S. Cloning of a novel ubiquitously expressed human phosphatidylinositol 3-kinase and identification of its binding site on p85. Mol. Cell. Biol. 13, 7677–7688 (1993).
Soltoff, S.P., Carraway, K.L., Prigent, S.A., Gullick, W.G. & Cantley, L.C. ErbB3 is involved in activation of phosphatidyinositol 3-kinase by epidermal growth factor. Mol. Cell. Biol. 14, 3550–3558 (1994).
Lev, S., Givol, D. & Yarden, Y. Interkinase domain of kit contains the binding site for phosphatidylinositol 3' kinase. Proc Natl. Acad. Sci. USA 89, 678–682 (1992).
Reedijk, M. et al. Tyr721 regulates specific binding of the CSF-1 receptor kinase insert to PI 3'-kinase SH2 domains: a model for SH2-mediated receptor-target interactions. Embo J. 11, 1365–1372 (1992).
Sun, X.J. et al. The structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature 352, 73–77 (1991).
Prasad, K.V.S. et al. T-cell antigen CD28 interacts with the lipid kinase phosphatidylinositol 3-kinase by a cytoplasmic pTyr-Met-Xxx-Met motif. Proc. Natl. Acad. Sci. USA 91, 2834–2838 (1994).
Auger, K.R., Carpenter, C.L., Shoelson, S.E., Piwnica-Worms, H. & Cantley, L.C. Polyoma virus middle T antigen/pp60c-src complex associates with purified phosphatidylinositol 3-kinase in vitro:dependence on protein-tyrosine kinase activity. J Biol. Chem. 267, 5408–5415 (1992).
Cantley, L.C. et al. Oncogenes and signal transduction. Cell 64, 281–302 (1991).
Songyang, Z. et al. SH2 domains recognize specific phosphopeptide sequences. Cell 72, 767–778 (1993).
Piccione, E. et al. PI 3-kinase p85 SH2 domains specificity defined by direct phosphopeptide/SH2 domain binding. Biochemistry 32, 3197–3202 (1993).
Songyang, Z. et al. Specific motifs recognized by the SH2 domains of Csk 3BP2, fps/fes, Grb-2, HCP, SHC, Syk and Vav. Mol. Cell. Biol. 14, 2777–2785 (1994).
Booker, G.W. et al. Structure of SH2 domain of the p85α subunit of phosphatidylinositol-3-OH kinase. Nature 358, 684–687 (1992).
Eck, M.J., Shoelson, S.E. & Harrison, S.C. Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature 362, 87–91 (1993).
Hatada, M.H. et al. Molecular basis for the interaction of the protein tyrosine kinase ZAP-70 with the T-cell receptor. Nature 377, 32–38 (1995).
Lee, C.H. et al. Crystal structures of peptide complexes of the N-terminal SH2 domain of the Syp tyrosine phosphatase. Structure 2, 423–438 (1994).
Pascal, S.M. et al. Nuclear magnetic resonance structure of an SH2 domain of PLC complexed with a high affinity binding peptide. Cell 77, 461–472 (1994).
Overduin, M., Rios, C.B., Mayer, B.J., Baltimore, D. & Cowburn, D. Three-Dimensional Solution Structure of the src Homology 2 Domain of c-abl. Cell 70, 697–704 (1992).
Waksman, G. et al. Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine phosphorylated peptides. Nature 356, 646–653 (1992).
Waksman, G., Shoelson, S.E., Pant, N., Cowburn, D. & Kuriyan, J. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: Crystal structures of the complexed and peptide-free forms. Cell 72, 779–790 (1993).
Zhou, M.M. et al. Solution structure of the She SH2 domain complexed with a tyrosine-phosphorylated peptide from the T-cell receptor. Proc. Natl. Acad. Sci. USA 92, 7784–7788 (1995).
Eck, M.J., Atwell, S.K., Shoelson, S.E. & Harrison, S.C. Crystal structure of the regulatory domains of the Src-family tyrosine kinase p56lck. Nature 368, 764–769 (1994).
Panayotou, G. et al. Interaction of the p85 subunit of PI 3-kinase and its N-terminal SH2 domain with a PDGF receptor phosphorylation site: structural features and analysis of conformational changes. Embo J. 11, 4261–4272 (1992).
Shoelson, S.E. et al. Specific phosphopeptide binding regulates a conformational change in the PI 3-kinase SH2 domain associated with enzyme activation. Embo J. 12, 795–802 (1993).
Williams, K.P. & Shoelson, S.E. A photoaffinity scan maps regions of the p85 SH2 domain involved in phosphoprotein binding. J. Biol. Chem. 268, 5362–5364 (1993).
Hensmann, M. et al. Phosphopeptide binding to the N-terminal SH2 domain of of the p85a subunit of PI 3-kinase: A heteronuclear NMR study. Prot. Sci. 3, 1020–1028 (1994).
Waksman, G. et al. Crystal structrue of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature 358, 646–653 (1992).
Eck, M.J., Pluskey, S., Trub, T., Harrison, S.C. & Shoelson, S.E. Spatial constraints on the recognition of phosphoproteins by the tandem SH2 domains of the phosphatase SH-PTP2. Nature 349, 277–280 (1996).
Songyang, Z., Gish, G., Mbamalu, G., Pawson, T. & Cantley, L.C. A single point mutation switches the specificity of group III Src homology (SH) 2 domains to that of group I SH2 domains. J. Biol. Chem. 270, 26029–26032 (1995).
Mahadevan, D. et al. Comparison of calcium-dependent co nformational changes in the N-terminal SH2 domains of p85 and GAP defines distinct properties for SH2 domains. Biochemistry 33, 746–754 (1994).
Backer, J.M. et al. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. Embo J. 11, 3469–3479 (1992).
Carpenter, C.L. et al. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85 kDa subunit. J. Biol. Chem. 268, 9478–9483 (1993).
Herbst, J.J. et al. Potent activation of phospahatidylinositol 3-kinase by simple phosphotyrosine peptides derived from insulin receptor substrate 1 containing two YMXM motifs for binding SH2 domains. Biochemistry 33, 9376–9381 (1994).
Marengere, L.E.M. et al. SH2 domain specificity and activity modified by a single residue. Nature 369, 502–505 (1994).
Sambrook, J., Fritsch, E. & Maniatis, T. Molecular Cloning: A laboratory manual (Cold Spring Harbor Laboratory Press, 1989).
Hendrickson, W.A., Horton, J.R. & LeMaster, D.M. Selenomethionyl proteins produced for analysis by multiwavelength anonomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9, 1665–1672 (1990).
Kabsch, W. Evaluation of single crystal diffraction data from a position sensitive detector. J. Appl. Crystallogr. 21, 916–924 (1988).
CollaborativeComputationalProjectNumber4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D50, 760–776 (1994).
Otwinowski, Z. Maximun likelihood refinement of heavy atom parameters (1991).
Cowtan, K. “DM”: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34–38 (1994).
Jones, T.A., Zou, J.Y., Cowan, S. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta. Crystallogr. A47, 110–119 (1991).
Brünger, A. X-PLOR Version 3.0: A System for Crystallography and NMR (Yale University Press, New Haven, 1992).
Navaza, J. . in Molecular Replacement: Proceedings of the CCP4 Study Weekend (eds. Dodson, E.J., Gover, S. & Wolf, W.) 87–90 (SERC, Daresbury, UK, 1992).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281–296 (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Nolte, R., Eck, M., Schlessinger, J. et al. Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat Struct Mol Biol 3, 364–374 (1996). https://doi.org/10.1038/nsb0496-364
Received:
Accepted:
Issue date:
DOI: https://doi.org/10.1038/nsb0496-364
This article is cited by
-
The kinetics of folding of the NSH2 domain from p85
Scientific Reports (2019)
-
Insights into the mechanism of the PIK3CA E545K activating mutation using MD simulations
Scientific Reports (2018)
-
Activation of PI3Kα by physiological effectors and by oncogenic mutations: structural and dynamic effects
Biophysical Reviews (2014)
-
Insights into structure and function of SHIP2-SH2: homology modeling, docking, and molecular dynamics study
Journal of Chemical Biology (2011)
-
Phosphotyrosine-binding domains in signal transduction
Nature Reviews Molecular Cell Biology (2002)