Abstract
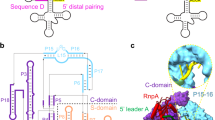
Guided by an in vitro selection experiment designed to obtain tight binding aptamers of Escherichia coli glutamine specific tRNA (tRNAGln) for glutaminyl-tRNA synthetase (GlnRS), we have engineered a tRNA mutant in which the five-nucleotide variable loop sequence 5′-44CAUUC48-3′ is replaced by 5′-44AGGU48-3′. This mutant tRNA binds to GlnRS with 30-fold improved affinity compared to the wild type. The 2.7 Å cocrystal structure of the RNA aptamer–GlnRS complex reveals major rearrangements in the central tertiary core of the tRNA, while maintaining an RNA–protein interface identical to the wild type. The repacked RNA core features a novel hydrogen bonding arrangement of the trans Levitt pair G15–U48, a new sulfate binding pocket in the major groove, and increased hydrophobic stacking interactions among the bases. These data suggest that enhanced protein binding to a mutant globular RNA can arise from stabilization of RNA tertiary interactions rather than optimization of RNA–protein contacts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Conn, G.L. & Draper, D.E. RNA structure. Curr. Opin. Struct. Biol. 8, 278–285 (1998).
Ferre-D'Amare, A.R. & Doudna, J.A. RNA folds: insights from recent crystal structures. Annu. Rev. Biophys. Biomol. Struct. 28, 57–73 (1999).
Cusack, S. RNA–protein complexes. Curr. Opin. Struct. Biol. 9, 66–73 (1999).
Rould, M.A., Perona, J.J., Soll, D. & Steitz, T.A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNAGln and ATP at 2.8 Å resolution. Science 246, 1135–1142 (1989).
Ruff, M. et al. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science 252, 1682–1689 (1991).
Biou, V., Yaremchuk, A., Tukalo, M. & Cusack, S. The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science 263, 1404–1410 (1994).
Cusack, S., Yaremchuk, A. & Tukalo, M. The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNALys and a T. thermophilus tRNALys transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J. 15, 6321–6334 (1996).
Cusack, S., Yaremchuk, A. & Krikliviy, I., Tukalo, M. tRNAPro anticodon recognition by Thermus thermophilus prolyl-tRNA synthetase. Structure 6, 101–108 (1998).
Silvian, L.F., Wang, J. & Steitz, T.A. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science 285, 1074–1077 (1999).
Sankaranarayanan, R. et al. The structure of threonyl-tRNA synthetase-tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell 97, 371–381 (1999).
Nissen, P. et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270, 1464–1472 (1995).
Nissen, P., Thirup, S., Kjeldgaard, M. & Nyborg, J. The crystal structure of Cys-tRNACys-EFTu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure 7, 143–156 (1999).
Schmitt, E., Panvert, M., Blanquet, S. & Mechulam, Y. Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet. EMBO J. 17, 6819–6826 (1998).
Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A. & Steinberg, S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26, 148–153 (1998).
Stubenrauch, M. In vitro selection and characterization of tRNA-like substrates of E. coli glutaminyl-tRNA synthetase. Doctoral thesis, Department of Chemistry and Biochemistry, University of Colorado, Boulder, Colorado; 1996.
Carey, J. Gel retardation. Methods Enzymol. 208, 103–117 (1991).
Rath, V.L., Silvian, L.F., Beijer, B., Sproat, B.S. & Steitz, T.A. How glutaminyl-tRNA synthetase selects glutamine. Structure, 6, 439–449 (1998).
Arnez, J.G. & Steitz, T.A. Crystal structure of unmodified tRNAGln complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33, 7560–7567 (1994).
Masquida, B., Sauter, C. & Westhof, E. A sulfate pocket formed by three G-U pairs in the 0.97 Å resolution X-ray structure of a nonameric RNA. RNA 5, 1384–1395 (1999).
Leontis, N.B. & Westhof, E. Conserved geometrical base-pairing patterns in RNA. Quat. Rev. Biophys. 31, 399–455 (1998).
Ferre-D'Amare, A.R., Zhou, K. & Doudna, J.A. Crystal structure of a hepatitis delta virus ribozyme. Nature 395, 567–574 (1998).
Convery, M.A. et al. Crystal structure of an RNA aptamer–protein complex at 2.8 Å resolution. Nature Struct. Biol. 5, 133–139 (1998).
Rowsell, S. et al. Crystal structures of a series of RNA aptamers complexed to the same protein target. Nature Struct. Biol. 5, 970–975 (1998).
Lim, W.A. & Sauer, R.T. Alternative packing arrangements in the hydrophobic core of λ repressor. Nature 339, 31–36 (1989).
Baldwin, E.P., Hajiseyedjavadi, O., Baase, W.A. & Matthews, B.W. The role of backbone flexibility in the accommodation of variants that repack the core of T4 lysozyme. Science 262, 1715–1718 (1993).
Lim, W.A., Hodel, A., Sauer, R.T. & Richards, F.M. The crystal structure of a mutant protein with altered but improved hydrophobic core packing. Proc. Natl. Acad. Sci. USA 91, 423–427 (1994).
Vetter, I.R. et al. Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme. Protein Sci. 5, 2399–2415 (1996).
Jen-Jacobson, L. Structural-perturbation approaches to thermodynamics of site-specific protein–DNA interactions. Methods Enzymol. 259, 305–344 (1995).
Jayaram, B., McConnell, K.J., Dixit, S.B. & Beveridge, D.L. Free energy analysis of protein–DNA binding: the EcoRI endonuclease–DNA complex. J. Comp. Phys. 151, 333–357 (1999).
Allain, F.H.-T., Gubser, C.C., Howe, P.W.A., Nagai, K., Neuhaus, D. & Varani, G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formation. Nature 380, 646–650 (1996).
Valegard, K., Murray, J.B., Stonehouse, N.J., van den Worm, S., Stockley, P.G. & Liljas, L. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein–RNA interactions. J. Mol. Biol. 270, 724–738 (1997).
Zidek, L., Novotny, M.V. & Stone, M.J. Increased protein backbone conformational entropy upon hydrophobic ligand binding. Nature Struct. Biol. 6, 1118–1121 (1999).
Grodberg, J. & Dunn, J.J. OmpT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J. Bacteriol. 170, 1245–1258 (1988).
Nissan, T.A., Oliphant, B. & Perona, J.J. (1999). An engineered class I transfer RNA with a class II tertiary fold. RNA 5, 434–445.
Milligan, J.F., Groebe, D.R., Witherell, G.W. & Uhlenbeck, O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 21, 8783–8798 (1987).
McGann, R.G. & Deutcher, M.P. Purification and characterization of a mutant tRNA nucleotidyl transferase. Eur. J. Biochem. 106, 321–328 (1980).
Hoben, P. et al. Escherichia coli glutaminyl-tRNA synthetase. J. Biol. Chem. 257, 11644–11650 (1982).
Perona, J.J., Swanson, R., Steitz, T.A. & Soll, D. Overproduction and purification of Escherichia coli tRNA2Gln and its use in crystallization of the glutaminyl-tRNA synthetase-tRNAGln complex. J. Mol. Biol. 202, 121–126 (1988).
Collaborative Computational Project Number 4, Acta Crystallogr. D 50, 760–776 (1994).
Tronrud, D.E., Ten Eyck, L.F. & Matthews, B.W. An efficient general-purpose least-squares refinement program for macromolecular structures Acta Crystallogr. A 43, 489–501 (1987).
Brunger, A.T., Kuriyan, J. & Karplus, M. Crystallographic R-factor refinement by molecular dynamics. Science 235, 458–460 (1987).
Jovine, L. et al. Crystal structure of the Ffh and EF-G binding sites in the conserved domain IV of E. coli 4.55 RNA. Structure 8, 527–540 (2000)
Acknowledgements
We thank O. Uhlenbeck and M. Derrick for encouragement and for communication of results prior to publication. We also thank B. Sproat and B. Beijer for the generous gift of QSI inhibitor. We are grateful to T. Earnest for assistance with data collection at ALS beamline 5.0.2, and to P. Allen for assistance with preparation of color figures. This work was supported by grants from the National Science Foundation (to J.J.P.) and from the Universitywide AIDS Research Program (to T.L.B.)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bullock, T., Sherlin, L. & Perona, J. Tertiary core rearrangements in a tight binding transfer RNA aptamer. Nat Struct Mol Biol 7, 497–504 (2000). https://doi.org/10.1038/75910
Received:
Accepted:
Issue date:
DOI: https://doi.org/10.1038/75910
This article is cited by
-
Post-SELEX optimization of aptamers
Analytical and Bioanalytical Chemistry (2016)
-
Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents
Molecular Therapy - Nucleic Acids (2014)
-
Study on an electrochemical biosensor for thrombin recognition based on aptamers and nano particles
Science in China Series B: Chemistry (2007)
-
Dpo4 is hindered in extending a G·T mismatch by a reverse wobble
Nature Structural & Molecular Biology (2004)