Abstract
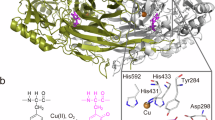
Mutation of six residues of Escherichia coli aspartate aminotransferase results in substantial acquisition of the transamination properties of tyrosine aminotransferase without loss of aspartate transaminase activity. X-ray crystallographic analysis of key inhibitor complexes of the hexamutant reveals the structural basis for this substrate selectivity. It appears that tyrosine aminotransferase achieves nearly equal affinities for a wide range of amino acids by an unusual conformational switch. An active-site arginine residue either shifts its position to electrostatically interact with charged substrates or moves aside to allow access of aromatic ligands.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Bocanegra, J.A., Scrutton, N.S. & Perham, R.N. Creation of an NADP-dependent pyruvate dehydrogenase multienzyme complex by protein engineering. Biochemistry. 32, 2737–2740. 1993.
Wilks, H.M. et al. Design of a specific phenyllactate dehydrogenase by peptide loop exchange on the Bacillus stearothermophilus lactate dehydrogenase framework. Biochemistry 31 7802–7806. 1992.
Wells, J.A., Cunningham, C., Graycer, T.P & Estell, D.A. Recruitment of substrate-specificity properties from one enzyme into a related one by protein engineering. Proc. natn. Acad. Sci. U.S.A. 84, 5167–5171. 1987.
Malcolm, B.A. & Kirsch, J.F. Site-directed mutagenesis of aspartate aminotransferase from Escherichia coli. Biochem. biophys. Res. Commun. 132, 915–921. 1985.
Kuramitsu, S., Okuno, S., Ogawa, T., Ogawa, H. & Kagamiyama, H. Aspartate aminotransferase of Escherichia coli: nucleotide sequence of the aspC gene. J. Biochem. (Tokyo) 97, 1259–1262. 1985.
Kuramitsu, S., Inoue, K., Ogawa, T., Ogawa, H. & Kagamiyama, H. Aromatic amino acid aminotransferase of Escherichia coli: nucleotide sequence of the tyrB gene. Biochem. biophys. Res. Commun. 133, 134–139. 1985.
Fotheringham, I.G. et al. The cloning and sequence analysis of the aspC and tyrB genes from Escherichia coli K12. Biochem. J. 234, 593–604. 1986.
Powell, J.T. & Morrison, J.F. The purification and properties of the aspartate aminotransferase and aromatic amino acid aminotransferase from Escherichia coli. Eur. J. Biochem. 87, 391–400. 1978.
Hayashi, H., Inoue, K., Nagata, T., Kuramitsu, S. & Kagamiyama, H. Escherichia coli aromatic amino acid aminotransferase: characterization and comparison with aspartate aminotransferase. Biochemistry. 32, 12229–12239. 1993.
Kirsch, J.F. & Onuffer, J.J. Redesign of aspartate aminotransferase specificity to that of tyrosine aminotransferase. in Biochemistry of Vitamin B6 and PQQ (eds, Bossa, F. et. al) 37–41. (Birkhäuser Verlag, Basel; 1994).
Jäger, J., Moser, M., Sauder, U. & Jansonius, J.N. Crystal structure of Escherichia coliaspartate aminotransferase in two conformations. Comparison of an unliganded open and two liganded closed forms. J. molec. Biol. 239, 285–305. 1994.
Seville, M., Vincent, M.G. & Hahn, K. Modeling the three-dimensional structures of bacterial aminotransferases. Biochemistry. 27, 8344–8349. 1988.
Jäger, J., Solmajer, T. & Jansonius, J.N. Computational approach towards the three-dimensional structure of Escherichia colityrosine aminotransferase. FEBS Letts. 306, 234–238. 1992.
Jansonius, J.N. & Vincent, M.G. Structural basis for catalysis of aspartate aminotransferase. in Biological Macromolecules and Assemblies, vol. 3 (eds. Jurnak, F.A. & McPherson, A.) 187–285 (Wiley, New York; 1987.)
Hayashi, H., Kuramitsu, S. & Kagamiyama, H. Replacement of an interdomain residue Val 39 of Escherichia coli aspartate aminotransferase affects the catalytic competence without altering the substrate specificity of the enzyme. J. Biochem (Tokyo). 109, 699–704. 1991.
Koehler, E. et al. Significant improvement to the catalytic properties of aspartate aminotransferase: role of hydrophobic and charged residues in the substrate binding pocket. Biochemistry. 33, 90–97. 1994.
Jäger, J., Pauptit, R.A., Sauder, U. & Jansonius, J.N. Three-dimensional structure of a mutant E. coli aspartate aminotransferase with increased enzymic activity. Prot. Engng. 7, 605–612. 1994.
Malashkevich, V.N., Toney, M.D. & Jansonius, J.N. Crystal structures of true enzymatic reaction intermediates: aspartate and glutamate ketimines in aspartate aminotransferase. Biochemistry. 32, 13451–13462. 1993.
Cronin, C.N. & Kirsch, J.F. Role of Arginine-292 in the substrate specificity of aspartate aminotransferase as examined by site-directed mutagenesis. Biochemistry. 27, 4572–4579. (1988).
Hayashi, H., Kuramitsu, S., Inoue, Y., Morino, Y. & Kagamiyama, H. [Arg 292 → Val] or [Arg 292 → Leu] mutation enhances the reactivity of Escherichia coliaspartate aminotransferase with aromatic amino acids. Biochem. biophys. Res. Commun. 159, 337–342. (1989).
Hedstrom, L., Szilagyi, L. & Rutter, W.J. Converting trypsin to chymotrypsin: the role of surface loops. Science 255, 1249–1253. (1992).
Messerschmidt, A. & Pflugrath, J.M. Crystal orientation and X-ray pattern prediction routines for area detector diffractometer. J. appl. Crystallogr. 20, 306–319. (1987).
Kabsch, W. Evaluation of single crystal X-ray diffraction from a position sensitive detector. J. appl. Crystallogr. 21, 916–924. (1988).
CCP4, SERC U.K.Collaborative Project (Daresbury Laboratory, U.K.; 1979.)
Jones, T.A. A graphic model building and refinement system for macromolecules. J. appl. Crystallogr. 11, 268–272. (1978).
Tronrud, D.E., Ten Eyck, L.F. & Matthews, B.W. An efficient general-purpose least-squares refinement program for macromolecular structures. Acta crystallogr. A43, 489–501. (1987).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta crystallogr A47, 110–119. (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Malashkevich, V., Onuffer, J., Kirsch, J. et al. Alternating arginine-modulated substrate specificity in an engineered tyrosine aminotransferase. Nat Struct Mol Biol 2, 548–553 (1995). https://doi.org/10.1038/nsb0795-548
Received:
Accepted:
Issue date:
DOI: https://doi.org/10.1038/nsb0795-548
This article is cited by
-
Computational remodeling of an enzyme conformational landscape for altered substrate selectivity
Nature Communications (2023)
-
Aspartate aminotransferase Rv3722c governs aspartate-dependent nitrogen metabolism in Mycobacterium tuberculosis
Nature Communications (2020)
-
Structural basis of substrate recognition by a novel thermostable (S)-enantioselective ω-transaminase from Thermomicrobium roseum
Scientific Reports (2019)
-
Crystal structures of Mycobacterium tuberculosis HspAT and ArAT reveal structural basis of their distinct substrate specificities
Scientific Reports (2016)
-
Molecular determinants for substrate selectivity of ω-transaminases
Applied Microbiology and Biotechnology (2012)