Abstract
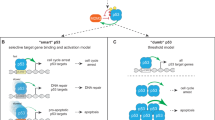
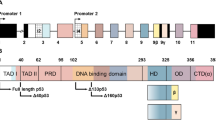
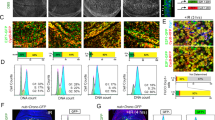
p53 is a nuclear phosphoprotein that regulates cellular fate after genotoxic stress through its role as a transcriptional regulator of genes involved in cell cycle control and apoptosis. The C-terminal region of p53 is known to negatively regulate sequence specific DNA-binding of p53; modifications to the C-terminus relieve this inhibition. Two models have been proposed to explain this latency: (i) an allosteric model in which the C-terminal domain interacts with another domain of p53 or (ii) a competitive model in which the C-terminal and the core domains compete for DNA binding. We have characterized latent and active forms of dimeric p53 using gel mobility shift assays and NMR spectroscopy. We show on the basis of chemical shifts that dimeric p53 both containing and lacking the C-terminal domain are identical in conformation and that the C-terminus does not interact with other p53 domains. Similarly, NMR spectra of isolated core and tetramerization domains confirm a modular p53 architecture. The data presented here rule out an allosteric model for the regulation of p53.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ko, L.J. & Prives, C. Genes Dev. 10, 1054–1072 (1996).
Levine, A.J. Cell 88, 323–331 (1997).
Kuerbitz S.J., Plunkett B.S., Walsh W.V. & Kastan M.B. Proc. Natl. Acad. Sci. USA 15, 7491–7495 (1992).
May, P. & May, E. Oncogene 18, 7621–7636 (1999).
Jayaraman, L. & Prives, C. Cell. Mol. Life Sci. 55, 76–87 (1999).
Halazonetis, T.D. & Kandil, A.N. EMBO J. 12, 5057–5064 (1993).
Hupp, T.R. & Lane, D.P. Curr. Biol. 4, 865–875 (1996).
El-Deiry, W.S., Kern, S.E., Pietenpol, J.A., Kinzler K.W. & Vogelstein, B. Nature Genet. 1, 45–49 (1992).
Funk, W.D., Pak, D.T., Karas, R.H., Wright, W.E. & Shay J.W. Mol. Cell Biol. 12, 2866–2871 (1992).
Hupp, T.R., Meek, D.W., Midgley, C.A. & Lane, D.P. Cell 71, 875–886 (1992).
Wolf, D., Harris, N., Goldfinger, N. & Rotter, V. Mol. Cell. Biol. 5, 127–132 (1985).
Wolkowicz R., Peled A., Elkind N.B. & Rotter V. Cancer Detect. Prev. 22, 1–13 (1998).
Zhao K., Chai X., Johnston K., Clements A. & Marmorstein R. J. Biol. Chem. 276, 12120–12127 (2001).
Cho, Y., Gorina, S., Jeffrey, P.D. & Pavletich, N.P. Science 265, 346–355 (1994).
Gorina, S. & Pavletich, N.P. Science 274, 1001–1005 (1996).
Lee, W. et al. Nature Struct. Biol. 1, 877–890 (1994).
Clore G.M. et al. Science 265, 386–391 (1994).
Jeffrey, R.D., Gorina, S. & Pavletich, N.P. Science 26, 1498–1502 (1995).
Kussie, P.H. et al. Science 274, 948–953 (1996).
Rustandi, R.R., Baldisseri, D.M. & Weber, D.J. Nature Struct. Biol. 7, 570–574 (2000).
Jayaraman J. & Prives, C. Cell 81, 1021–1029 (1995).
Davison, T.S. et al. J. Mol. Biol. 307, 605–617 (2001).
Matsumura, I. & Ellington, A.D. Protein Sci. 8, 731–740 (1999).
Anderson, M.E., Woelker, B., Reed, M., Wang, P. & Tegtmeyer, P. Mol. Cell. Biol. 17, 6255–6264 (1997).
Mclure, K.G. & Lee, P.W.K. EMBO J. 17, 3342–3350 (1998).
Mulder, F.A.A., Schipper, D., Bott, R. & Boelens, R. J. Mol. Biol. 292, 111–123 (1999).
Seavey, B.R., Farr, E.A., Westler, W.M. & Markley, J.L. J. Biomol. NMR 1, 217–236 (1991).
Pavletich, N.P, Chambers, K.A. & Pabo, C.O. Genes Dev. 7, 2556–2564 (1993).
Metzler, W.J. et al. Biochemistry 32, 13818–13829 (1993).
Wishart, D.S. & Sykes, B.D. Methods Enzymol. 239, 363–392 (1994).
Gardner, K.H. & Kay, L.E. Annu. Rev. Biophys. Biomol. Struct. 27, 357–406 (1998).
Delaglio, F. et al. J. Biomol. NMR 6, 277–293 (1995).
Pervushin, K., Riek, R., Wider, G. & Wüthrich, K. Proc. Natl. Acad. Sci. USA 94, 12366–12371 (1997).
Yang, D. & Kay, L.E. J. Am. Chem. Soc. 121, 2571–2575 (1999).
Konrat, R., Yang, D. & Kay, L.E. J. Biomol. NMR 15, 309–313 (1999).
Mulder, F.A.A., Ayed, A., Yang, D., Arrowsmith, C.H. & Kay, L.E. J. Biomol. NMR 18, 173–176 (2000).
Acknowledgements
We thank A. Pineda-Lucena and T. Davison for useful discussions. This work was supported by the National Cancer Institute of Canada with funds from the Canadian Cancer Society and by the Canadian Institutes of Health Research (CIHR). A.A. is the recipient of the Governor General's award for Leukemia Research from the Leukemia Research Fund of Canada. F.A.A.M. is the recipient of a postdoctoral fellowship from the European molecular Biology Organization. L.E.K. is a foreign investigator of the Howard Hughes Medical Research Institute. C.H.A. is a CIHR scientist.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ayed, A., Mulder, F., Yi, GS. et al. Latent and active p53 are identical in conformation. Nat Struct Mol Biol 8, 756–760 (2001). https://doi.org/10.1038/nsb0901-756
Received:
Accepted:
Issue date:
DOI: https://doi.org/10.1038/nsb0901-756
This article is cited by
-
Intracellular FGF1 protects cells from apoptosis through direct interaction with p53
Cellular and Molecular Life Sciences (2023)
-
Membrane permeabilization is mediated by distinct epitopes in mouse and human orthologs of the necroptosis effector, MLKL
Cell Death & Differentiation (2022)
-
P53 aggregation, interactions with tau, and impaired DNA damage response in Alzheimer’s disease
Acta Neuropathologica Communications (2020)
-
ODiNPred: comprehensive prediction of protein order and disorder
Scientific Reports (2020)
-
The multiple mechanisms that regulate p53 activity and cell fate
Nature Reviews Molecular Cell Biology (2019)