Abstract
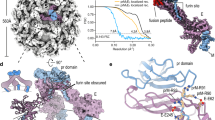
The current Zika virus (ZIKV) epidemic is characterized by severe pathogenicity in both children and adults. Sequence changes in ZIKV since its first isolation are apparent when pre-epidemic strains are compared with those causing the current epidemic. However, the residues that are responsible for ZIKV pathogenicity are largely unknown. Here we report the cryo-electron microscopy (cryo-EM) structure of the immature ZIKV at 9-Å resolution. The cryo-EM map was fitted with the crystal structures of the precursor membrane and envelope glycoproteins and was shown to be similar to the structures of other known immature flaviviruses. However, the immature ZIKV contains a partially ordered capsid protein shell that is less prominent in other immature flaviviruses. Furthermore, six amino acids near the interface between pr domains at the top of the spikes were found to be different between the pre-epidemic and epidemic ZIKV, possibly influencing the composition and structure of the resulting viruses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Schuler-Faccini, L. et al. Possible association between Zika virus infection and microcephaly - Brazil, 2015. MMWR Morb. Mortal. Wkly. Rep. 65, 59–62 (2016).
Chan, J.F., Choi, G.K., Yip, C.C., Cheng, V.C. & Yuen, K.Y. Zika fever and congenital Zika syndrome: An unexpected emerging arboviral disease. J. Infect. 72, 507–524 (2016).
Lindenbach, B.D., Murray, C.L., Thiel, H.-J. & Rice, C.M. Fields Virology (eds. Knipe, D.M. et al.) (Lippincott Williams & Wilkins, 2013).
Mukhopadhyay, S., Kuhn, R.J. & Rossmann, M.G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3, 13–22 (2005).
Kuhn, R.J. et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108, 717–725 (2002).
Mukhopadhyay, S., Kim, B.S., Chipman, P.R., Rossmann, M.G. & Kuhn, R.J. Structure of West Nile virus. Science 302, 248 (2003).
Zhang, X. et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 20, 105–110 (2013).
Sirohi, D. et al. The 3.8 Å resolution cryo-EM structure of Zika virus. Science 352, 467–470 (2016).
Kostyuchenko, V.A. et al. Structure of the thermally stable Zika virus. Nature 533, 425–428 (2016).
Zhang, Y. et al. Structures of immature flavivirus particles. EMBO J. 22, 2604–2613 (2003).
Stadler, K., Allison, S.L., Schalich, J. & Heinz, F.X. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71, 8475–8481 (1997).
Yu, I.-M. et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319, 1834–1837 (2008).
Rodenhuis-Zybert, I.A. et al. Immature dengue virus: a veiled pathogen? PLoS Pathog. 6, e1000718 (2010).
Colpitts, T.M. et al. prM-antibody renders immature West Nile virus infectious in vivo. J. Gen. Virol. 92, 2281–2285 (2011).
Li, L. et al. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319, 1830–1834 (2008).
Kostyuchenko, V.A., Zhang, Q., Tan, J.L., Ng, T.S. & Lok, S.M. Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J. Virol. 87, 7700–7707 (2013).
Ma, L., Jones, C.T., Groesch, T.D., Kuhn, R.J. & Post, C.B. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl. Acad. Sci. USA 101, 3414–3419 (2004).
Dokland, T. et al. West Nile virus core protein; tetramer structure and ribbon formation. Structure 12, 1157–1163 (2004).
Wang, L. et al. From mosquitos to humans: genetic evolution of Zika virus. Cell Host Microbe 19, 561–565 (2016).
Pierson, T.C. et al. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe 1, 135–145 (2007).
Nelson, S. et al. Maturation of West Nile virus modulates sensitivity to antibody-mediated neutralization. PLoS Pathog. 4, e1000060 (2008).
Davenport, T.M. et al. Isolate-specific differences in the conformational dynamics and antigenicity of HIV-1 gp120. J. Virol. 87, 10855–10873 (2013).
Kwong, P.D. et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 (2002).
Dejnirattisai, W. et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat. Immunol. 17, 1102–1108 (2016).
Tirado, S.M. & Yoon, K.J. Antibody-dependent enhancement of virus infection and disease. Viral Immunol. 16, 69–86 (2003).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Scheres, S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Guo, F. & Jiang, W. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol. Biol. 1117, 401–443 (2014).
Pettersen, E.F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Katoh, K. & Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Pickett, B.E. et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 40, D593–D598 (2012).
Acknowledgements
We thank M. Sevvana for discussions about the manuscript. We thank S. Kelly for helping us prepare the manuscript. We also thank the Purdue Cryo-EM Facility for equipment access and support. This work was supported by the National Institutes of Health (RO1 AI076331 to M.G.R. and R.J.K., and a subaward for RO1 AI073755 (principal investigator: M.S. Diamond, Washington University) to both M.G.R. and R.J.K.).
Author information
Authors and Affiliations
Contributions
A.S.M., D.S. and G.B. were involved in preparation of cell culture, and optimization and purification of virus sample; V.M.P. and T.K. conducted the cryo-EM preparation, data collection and data processing; V.M.P. performed the data analyses; W.J. made his JSPR program available for reconstruction and refinement of the cryo-EM map; and V.M.P., R.J.K. and M.G.R. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Resolution estimation using Fourier shell correlation (FSC).
A plot of FSC against resolution in Å-1, calculated using two independent half-sets of the cryo-EM data. The resolution of the map is 9.1 Å using the 0.143 FSC cut-off (light blue line).
Supplementary Figure 2 Fit of the DENV-2 prM–E crystal structure into the immature ZIKV cryo-EM map.
a, Surface view of the complete immature ZIKV. One of the 60 trimeric spikes is identified in color, with its E proteins in green and the pr domains in orange. The asymmetric unit is shown as a black triangle. The scale bar is 100 Å long. b, Top view showing the fit of three prM-E heterodimers into the cryo-EM spike density. The three independent prM-E heterodimers are colored in light green, blue and orange for the E proteins and dark green, dark blue and red for the pr domains, respectively. c, Side-view of the fitted prM-E heterodimers in a spike. The dashed black line shows the position of the viral membrane surface. d, Cryo-EM density showing the individual fitting of the transmembrane regions of E (blue) and M (magenta) proteins. Scale bar is 50 Å long in b, c and d.
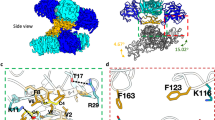
Supplementary Figure 3 Analysis of crystal structures fitted into immature ZIKV cryo-EM map.
a, Cryo-EM density showing the conservation of Asn-69 glycosylation site in the pr domains of immature ZIKV. The DENV-2 prM-E crystal structure (PDB ID: 3C6E) including the Asn-69 glycan has been fitted into all three positions within a trimeric spike. The glycans attached to Asn-69 are shown in blue. Scale bar is 25 Å in length. b, Glycan (colored in blue) attached to Asn-67 in the DENV-2 E protein structure and fitted into the map of the immature ZIKV showing the absence of density associated with the glycan at this site in ZIKV. c, Additional density adjacent to Asn-154 (red) in immature ZIKV indicating the presence of a glycan. Scale bar is 10 Å long in b and c. Black arrows in a and c point to the densities of the glycans. The E proteins and pr domains are shown in green and orange respectively in all panels.
Supplementary Figure 4 Alignment of sequences that form the interfaces between immature ZIKV spikes.
Top panel, Alignment of pr domain sequences from different flaviviruses. The residues that differ between the pre-epidemic (African) and epidemic (Asian) ZIKV strains are indicated in blue boxes. Bottom panel, Alignment of parts of the E protein sequence from different flaviviruses. In both panels, red columns indicate strictly conserved residues, yellow columns show partially conserved residues and white columns show variable residues. Residues involved in interactions within a trimeric spike and between adjacent spikes are indicated by brown and green colored bars above the corresponding residues, respectively. The furin cleavage site at the end of the pr domain is shown by a grey bar. Abbreviations DEN, JEV and YFV refer to dengue virus, Japanese encephalitis virus and yellow fever virus respectively.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Table 1 (PDF 1060 kb)
Rights and permissions
About this article
Cite this article
Prasad, V., Miller, A., Klose, T. et al. Structure of the immature Zika virus at 9 Å resolution. Nat Struct Mol Biol 24, 184–186 (2017). https://doi.org/10.1038/nsmb.3352
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/nsmb.3352
This article is cited by
-
Irreversible furin cleavage site exposure renders immature tick-borne flaviviruses fully infectious
Nature Communications (2025)
-
Identification of candidate genes involved in Zika virus-induced reversible paralysis of mice
Scientific Reports (2025)
-
Zika virus prM protein contains cholesterol binding motifs required for virus entry and assembly
Nature Communications (2023)
-
Zika virus M protein latches and locks the E protein from transitioning to an immature state after prM cleavage
npj Viruses (2023)
-
Adaptation to host cell environment during experimental evolution of Zika virus
Communications Biology (2022)