Abstract
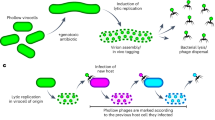
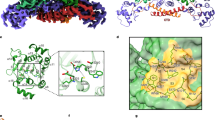
Prolyl cis-trans isomerizations are intrinsically slow reactions and known to be rate-limiting in many protein folding reactions. Here we report that a proline is used as a molecular timer in the infection of Escherichia coli cells by the filamentous phage fd. The phage is activated for infection by the disassembly of the two N-terminal domains, N1 and N2, of its gene-3-protein, which is located at the phage tip. Pro213, in the hinge between N1 and N2, sets a timer for the infective state. The timer is switched on by cis-to-trans and switched off by the unusually slow trans-to-cis isomerization of the Gln212-Pro213 peptide bond. The switching rate and thus the infectivity of the phage are determined by the local sequence around Pro213, and can be tuned by mutagenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Grathwohl, C. & Wüthrich, K. NMR studies of the rates of proline cis-trans isomerization in oligopeptides. Biopolymers 20, 2623–2633 (1981).
Stein, R.L. Mechanism of enzymatic and nonenzymatic prolyl cis-trans isomerization. Adv. Protein Chem. 44, 1–24 (1993).
Fischer, G. Chemical aspects of peptide bond isomerisation. Chem. Soc. Rev. 29, 119–127 (2000).
Reimer, U. et al. Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J. Mol. Biol. 279, 449–460 (1998).
Brandts, J.F., Halvorson, H.R. & Brennan, M. Consideration of the possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry 14, 4953–4963 (1975).
Balbach, J. & Schmid, F.X. In Mechanisms of Protein Folding (ed. Pain, R.H.) 212–237 (Oxford Univ. Press, Oxford, UK, 2000).
Kubelka, J., Eaton, W.A. & Hofrichter, J. Experimental tests of villin subdomain folding simulations. J. Mol. Biol. 329, 625–630 (2003).
Myers, J.K. & Oas, T.G. Mechanisms of fast protein folding. Annu. Rev. Biochem. 71, 783–815 (2002).
Schmid, F.X. Prolyl isomerases. Adv. Protein Chem. 59, 243–282 (2001).
Yaffe, M.B. et al. Sequence-specific and phosphorylation-dependent proline isomerization—a potential mitotic regulatory mechanism. Science 278, 1957–1960 (1997).
Fischer, G. & Aumüller, T. Regulation of peptide bond cis/trans isomerization by enzyme catalysis and its implication in physiological processes. Rev. Physiol. Biochem. Pharmacol. 148, 105–150 (2004).
Lubkowski, J., Hennecke, F., Plückthun, A. & Wlodawer, A. The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of G3P. Nat. Struct. Biol. 5, 140–147 (1998).
Holliger, P., Riechmann, L. & Williams, R.L. Crystal structure of the two N-terminal domains of g3p from filamentous phage fd at 1.9 angström: evidence for conformational lability. J. Mol. Biol. 288, 649–657 (1999).
Chatellier, J. et al. Interdomain interactions within the gene 3 protein of filamentous phage. FEBS Lett. 463, 371–374 (1999).
Karlsson, F., Borrebaeck, C.A., Nilsson, N. & Malmborg-Hager, A.C. The mechanism of bacterial infection by filamentous phages involves molecular interactions between TolA and phage protein 3 domains. J. Bacteriol. 185, 2628–2634 (2003).
Deng, L.W. & Perham, R.N. Delineating the site of interaction on the pIII protein of filamentous bacteriophage fd with the F-pilus of Escherichia coli. J. Mol. Biol. 319, 603–614 (2002).
Click, E.M. & Webster, R.E. Filamentous phage infection: required interactions with the TolA protein. J. Bacteriol. 179, 6464–6471 (1997).
Lubkowski, J., Hennecke, F., Plückthun, A. & Wlodawer, A. Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Structure 7, 711–722 (1999).
Stengele, I., Bross, P., Garces, X., Giray, J. & Rasched, I. Dissection of functional domains in phage fd adsorption protein. Discrimination between attachment and penetration sites. J. Mol. Biol. 212, 143–149 (1990).
Riechmann, L. & Holliger, P. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of. E. coli. Cell 90, 351–360 (1997).
Martin, A. & Schmid, F.X. The folding mechanism of a two-domain protein: folding kinetics and domain docking of the gene-3-protein of phage fd. J. Mol. Biol. 329, 599–610 (2003).
Martin, A. & Schmid, F.X. A proline switch controls folding and domain interactions in the gene-3-protein of the filamentous phage fd. J. Mol. Biol. 331, 1131–1140 (2003).
Frost, L.S. In Bacterial Conjugation (ed. Clewell, D.B.) 189–221 (Plenum Press, New York, 1993).
Martin, A. & Schmid, F.X. Evolutionary stabilization of the gene-3-protein of phage fd reveals the principles that govern the thermodynamic stability of two-domain proteins. J. Mol. Biol. 328, 863–875 (2003).
Lu, K.P., Hanes, S.D. & Hunter, T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380, 544–547 (1996).
Andreotti, A.H. Native state proline isomerization: An intrinsic molecular switch. Biochemistry 42, 9515–9524 (2003).
Weiwad, M. et al. Catalysis of proline-directed protein phosphorylation by peptidyl-prolyl cis/trans isomerases. J. Mol. Biol. 339, 635–646 (2004).
Lopez-Ilasaca, M. et al. Effects of FK506-binding protein 12 and FK506 on autophosphorylation of epidermal growth factor receptor. J. Biol. Chem. 273, 9430–9434 (1998).
Mallis, R.J., Brazin, K.N., Fulton, D.B. & Andreotti, A.H. Structural characterization of a proline-driven conformational switch within the Itk SH2 domain. Nat. Struct. Biol. 9, 900–905 (2002).
Gitti, R.K. et al. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273, 231–235 (1996).
Krebber, C. et al. Selectively-infective phage (SIP): a mechanistic dissection of a novel in vivo selection for protein-ligand interactions. J. Mol. Biol. 268, 607–618 (1997).
Sieber, V., Plückthun, A. & Schmid, F.X. Selecting proteins with improved stability by a phage-based method. Nat. Biotechnol. 16, 955–960 (1998).
Koradi, R., Billeter, M. & Wüthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 (1996).
Acknowledgements
We thank C. Unverzagt for help with peptide synthesis, M. Zeeb for help with NMR measurements, G. Fischer for a sample of cyclophilin 18, P. Holliger for E. coli HB2156 and C. Lehner, W. Schumann, B. Westermann and the members of our group for comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Additional competitive infections. (PDF 143 kb)
Supplementary Fig. 2
Binding of TolA to N1. (PDF 181 kb)
Rights and permissions
About this article
Cite this article
Eckert, B., Martin, A., Balbach, J. et al. Prolyl isomerization as a molecular timer in phage infection. Nat Struct Mol Biol 12, 619–623 (2005). https://doi.org/10.1038/nsmb946
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/nsmb946
This article is cited by
-
Genome-wide characterization of peptidyl-prolyl cis–trans isomerases in Penicillium and their regulation by salt stress in a halotolerant P. oxalicum
Scientific Reports (2021)
-
Genome organisation and comparative genomics of four novel Wolbachia genome assemblies from Indian Drosophila host
Functional & Integrative Genomics (2019)
-
Initiation of prolyl cis-trans isomerisation in the CDR-H3 loop of an antibody in response to antigen binding
Scientific Reports (2017)
-
Cis–trans peptide variations in structurally similar proteins
Amino Acids (2012)
-
The cytoplasmic cyclophilin from Azotobacter vinelandii interacts with phosphate acetyltransferase isoforms enhancing their in vitro activity
Molecular Biology Reports (2012)