Abstract
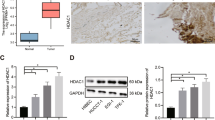
Expression and cellular distribution of claudin-1, a tight junction protein, is dysregulated in colon cancer and its overexpression in colon cancer cells induced dedifferentiation and increased invasion. However, the molecular mechanism(s) underlying dysregulated claudin-1 expression in colon cancer remains poorly understood. Histone deacetylase (HDAC)-dependent histone acetylation is an important mechanism of the regulation of cancer-related genes and inhibition of HDACs induces epithelial differentiation and decreased invasion. Therefore, in this study, we examined the role of HDAC-dependent epigenetic regulation of claudin-1 in colon cancer. In this study, we show that sodium butyrate and Trichostatin A (TSA), two structurally different and widely used HDAC inhibitors, inhibited claudin-1 expression in multiple colon cancer cell lines. Further studies revealed modulation of claudin-1 mRNA stability by its 3′-UTR as the major mechanism underlying HDAC-dependent claudin-1 expression. In addition, overexpression of claudin-1 abrogated the TSA-induced inhibition of invasion in colon cancer cells suggesting functional crosstalk. Analysis of mRNA expression in colon cancer patients, showed a similar pattern of increase in claudin-1 and HDAC-2 mRNA expression throughout all stages of colon cancer. Inhibition of claudin-1 expression by HDAC-2-specific small interfering RNA further supported the role of HDAC-2 in this regulation. Taken together, we report a novel post-transcriptional regulation of claudin-1 expression in colon cancer cells and further show a functional correlation between claudin-1 expression and TSA-mediated regulation of invasion. As HDAC inhibitors are considered to be promising anticancer drugs, these new findings will have implications in both laboratory and clinical settings.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Abuazza G, Becker A, Williams SS, Chakravarty S, Truong HT, Lin F et al. (2006). Claudins 6, 9, and 13 are developmentally expressed renal tight junction proteins. Am J Physiol Renal Physiol 291: F1132–F1141.
Chavey C, Muhlbauer M, Bossard C, Freund A, Durand S, Jorgensen C et al. (2008). Interleukin-8 expression is regulated by histone deacetylases through the NF-{kappa}B pathway in breast cancer. Mol Pharmacol 74: 1359–1366.
Dhawan P, Singh AB, Deane NG, No Y, Shiou S-R, Schmidt C et al. (2005). Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest 115: 1765–1776.
Dokmanovic M, Marks PA . (2005). Prospects: histone deacetylase inhibitors. J Cell Biochem 96: 293–304.
Godman CA, Joshi R, Tierney BR, Greenspan E, Rasmussen TP, Wang HW et al. (2008). HDAC3 impacts multiple oncogenic pathways in colon cancer cells with effects on Wnt and vitamin D signaling. Cancer Biol Ther 7: 1570–1580.
Honda H, Pazin MJ, D’Souza T, Ji H, Morin PJ . (2007). Regulation of the CLDN3 gene in ovarian cancer cells. Cancer Biol Ther 6: 1733–1742.
Honda H, Pazin MJ, Ji H, Wernyj RP, Morin PJ . (2006). Crucial roles of Sp1 and epigenetic modifications in the regulation of the CLDN4 promoter in ovarian cancer cells. J Biol Chem 281: 21433–21444.
Liu P-Y, Chan JY-H, Lin H-C, Wang S-L, Liu S-T, Ho C-L et al. (2008). Modulation of the cyclin-dependent kinase inhibitor p21WAF1/Cip1 gene by Zac1 through the antagonistic regulators p53 and histone deacetylase 1 in HeLa cells. Mol Cancer Res 6: 1204–1214.
Minucci S, Pelicci PG . (2006). Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 6: 38–51.
Nishikiori N, Sawada N, Ohguro H . (2008). Prevention of murine experimental corneal trauma by epigenetic events regulating claudin 6 and claudin 9. Jpn J Ophthalmol 52: 195–203.
Osanai M, Murata M, Chiba H, Kojima T, Sawada N . (2007a). Epigenetic silencing of claudin-6 promotes anchorage-independent growth of breast carcinoma cells. Cancer Sci 98: 1557–1562.
Osanai M, Murata M, Nishikiori N, Chiba H, Kojima T, Sawada N . (2007b). Occludin-mediated premature senescence is a fail-safe mechanism against tumorigenesis in breast carcinoma cells. Cancer Sci 98: 1027–1034.
Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S . (2001). Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276: 73–81.
Pryzbylkowski P, Obajimi O, Keen J . (2008). Trichostatin A and 5 Aza-2′ deoxycytidine decrease estrogen receptor mRNA stability in ER positive MCF7 cells through modulation of HuR. Breast Cancer Res Treat 111: 15–25.
Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H . (2006). Quantitative real-time RT-PCR data analysis: current concepts and the novel ‘gene expression's CT difference’ formula. J Mol Med 84: 901–910.
Shiou SR, Singh AB, Moorthy K, Datta PK, Washington MK, Beauchamp RD et al. (2007). Smad4 regulates claudin-1 expression in a transforming growth factor-beta-independent manner in colon cancer cells. Cancer Res 67: 1571–1579.
Wu Y, Starzinski-Powitz A, Guo S-W . (2007). Trichostatin A, a histone deacetylase inhibitor, attenuates invasiveness and reactivates E-cadherin expression in immortalized endometriotic cells. Reprod Sci 14: 374–382.
Yamamichi N, Yamamichi-Nishina M, Mizutani T, Watanabe H, Minoguchi S, Kobayashi N et al. (2005). The Brm gene suppressed at the post-transcriptional level in various human cell lines is inducible by transient HDAC inhibitor treatment, which exhibits antioncogenic potential. Oncogene 24: 5471–5481.
Zhang Z, Sheng H, Shao J, Beauchamp RD, DuBois RN . (2000). Posttranscriptional regulation of cyclooxygenase-2 in rat intestinal epithelial cells. Neoplasia 2: 523–530.
Zhu P, Martin E, Mengwasser J, Schlag P, Janssen K-P, Göttlicher M . (2004). Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell 5: 455–463.
Acknowledgements
This work was supported by NIH grant CA119005, CA124977 (P Dhawan), and AHA Grant 0435471N, 5P50DK044757 and P30DK058406 Pilot project (AB Singh), the Society of University Surgeons-Ethicon Scholarship Award (JJS) CA112215 (TJ Yeatman), DK052334, CA 069457, the GI Cancer SPORE grant CA95103 (RD Beauchamp), the Vanderbilt-Ingram Cancer Center P30CA68485 and the NIH grant supporting the Digestive Diseases Center DK58404. We thank Christian Kis for the help in real-time quantitative reverse transcription–PCR analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Krishnan, M., Singh, A., Smith, J. et al. HDAC inhibitors regulate claudin-1 expression in colon cancer cells through modulation of mRNA stability. Oncogene 29, 305–312 (2010). https://doi.org/10.1038/onc.2009.324
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/onc.2009.324
Keywords
This article is cited by
-
The role of epigenetic modifications in Colorectal Cancer Metastasis
Clinical & Experimental Metastasis (2022)
-
Loss of claudin-3 expression induces IL6/gp130/Stat3 signaling to promote colon cancer malignancy by hyperactivating Wnt/β-catenin signaling
Oncogene (2017)
-
Claudins in cancer: bench to bedside
Pflügers Archiv - European Journal of Physiology (2017)
-
Regulatory dissection of the CBX5 and hnRNPA1 bi-directional promoter in human breast cancer cells reveals novel transcript variants differentially associated with HP1α down-regulation in metastatic cells
BMC Cancer (2016)
-
The effects of a novel aliphatic-chain hydroxamate derivative WMJ-S-001 in HCT116 colorectal cancer cell death
Scientific Reports (2015)