Abstract
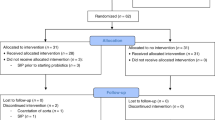
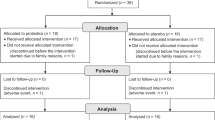
The fecal microbiota of 37 infants with (n = 20) or without (n = 17) probiotic administration was evaluated on D 3, and at 1, 3, and 12 mo by fluorescence in situ hybridization-flow cytometry (FISH-FC), PCR, and bacteriological culture methods. They represent consecutive subjects of an ongoing double-blind, placebo-controlled trial on a probiotic formula (LGG and Bifidobacterium longum) administered during the first 6 mo of life. Despite varying composition in each baby, there was a general bacterial colonization pattern in the first year. Bifidobacteria increased markedly (p = 0.0003) with a parallel decrease in Enterobacteriaceae (p < 0.001) and Bacteroides–Prevotella (p = 0.005) populations. Eubacterium rectale–Clostridium coccoides (p < 0.001) and Atopobium (p = 0.039) groups also gradually increased. This overall pattern was unaffected by probiotic administration (p > 0.05). B. longum (p = 0.005) and Lactobacillus rhamnosus (p < 0.001) were detected more frequently in probiotic group during supplementation, but no difference after supplementation had ceased (p > 0.05). Cultured lactic acid bacteria were also more numerous in the probiotic-administered babies during treatment period (log10 CFU/g 8.4 versus 7.4; p = 0.035). Our results indicate that supplemented strains could be detected but did not persist in the bowel once probiotic administration had ceased.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Abbreviations
- LAB:
-
lactic acid bacteria
References
Gillman MW 2005 Developmental origins of health and disease. N Engl J Med 353: 1848–1850
Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE 2006 Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118: 511–521
Hooper LV, Gordon JI 2001 Commensal host-bacterial relationships in the gut. Science 292: 1115–1118
Tannock GW 2005 Commentary: remembrance of microbes past. Int J Epidemiol 34: 13–15
Langendijk PS, Schut F, Jansen GJ, Raangs GC, Kamphuis GR, Wilkinson MH, Welling GW 1995 Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol 61: 3069–3075
Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Dore J 1999 Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 65: 4799–4807
Wilson KH, Blitchington RB 1996 Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol 62: 2273–2278
Lay C, Rigottier-Gois L, Holmstrom K, Rajilic M, Vaughan EE, de Vos WM, Collins MD, Thiel R, Namsolleck P, Blaut M, Dore J 2005 Colonic microbiota signatures across five northern European countries. Appl Environ Microbiol 71: 4153–4155
Lay C, Sutren M, Rochet V, Saunier K, Dore J, Rigottier-Gois L 2005 Design and validation of 16S rRNA probes to enumerate members of the Clostridium leptum subgroup in human faecal microbiota. Environ Microbiol 7: 933–946
Strachan DP 2000 Family size, infection and atopy: the first decade of the “hygiene hypothesis.”. Thorax 55: S2–S10
Bjorksten B, Sepp E, Julge K, Voor T, Mikelsaar M 2001 Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol 108: 516–520
Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E 2001 Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357: 1076–1079
Reid G, Sanders ME, Gaskins HR, Gibson GR, Mercenier A, Rastall R, Roberfroid M, Rowland I, Cherbut C, Klaenhammer TR 2003 New scientific paradigms for probiotics and prebiotics. J Clin Gastroenterol 37: 105–118
Garrido D, Suau A, Pochart P, Cruchet S, Gotteland M 2005 Modulation of the fecal microbiota by the intake of a Lactobacillus johnsonii La1-containing product in human volunteers. FEMS Microbiol Lett 248: 249–256
Rochet V, Rigottier-Gois L, Sutren M, Krementscki MN, Andrieux C, Furet JP, Tailliez P, Levenez F, Mogenet A, Bresson JL, Meance S, Cayuela C, Leplingard A, Dore J 2006 Effects of orally administered Lactobacillus casei DN-114 001 on the composition or activities of the dominant faecal microbiota in healthy humans. Br J Nutr 95: 421–429
Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP 2001 Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol 67: 2578–2585
Yamano T, Iino H, Takada M, Blum S, Rochat F, Fukushima Y 2006 Improvement of the human intestinal flora by ingestion of the probiotic strain Lactobacillus johnsonii La1. Br J Nutr 95: 303–312
Chew FT, Lim SH, Goh DY, Lee BW 1999 Sensitization to local dust-mite fauna in Singapore. Allergy 54: 1150–1159
Knarreborg A, Simon MA, Engberg RM, Jensen BB, Tannock GW 2002 Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl Environ Microbiol 68: 5918–5924
Langhendries JP, Detry J, Van Hees J, Lamboray JM, Darimont J, Mozin MJ, Secretin MC, Senterre J 1995 Effect of a fermented infant formula containing viable bifidobacteria on the fecal flora composition and pH of healthy full-term infants. J Pediatr Gastroenterol Nutr 21: 177–181
Kwon HS, Yang EH, Lee SH, Yeon SW, Kang BH, Kim TY 2005 Rapid identification of potentially probiotic Bifidobacterium species by multiplex PCR using species-specific primers based on the region extending from 16S rRNA through 23S rRNA. FEMS Microbiol Lett 250: 55–62
Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H 1999 Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol 65: 4506–4512
Brandt K, Alatossava T 2003 Specific identification of certain probiotic Lactobacillus rhamnosus strains with PCR primers based on phage-related sequences. Int J Food Microbiol 84: 189–196
Mah KW, Bjorksten B, Lee BW, van Bever HP, Shek LP, Tan TN, Lee YK, Chua KY 2006 Distinct pattern of commensal gut microbiota in toddlers with eczema. Int Arch Allergy Immunol 140: 157–163
Gueimonde M, Kalliomaki M, Isolauri E, Salminen S 2006 Probiotic intervention in neonates—will permanent colonization ensue?. J Pediatr Gastroenterol Nutr 42: 604–606
Hayashi H, Sakamoto M, Kitahara M, Benno Y 2006 Diversity of the Clostridium coccoides group in human fecal microbiota as determined by 16S rRNA gene library. FEMS Microbiol Lett 257: 202–207
Kukkonen K, Savilahti E, Haahtela T, Juntunen-Backman K, Korpela R, Poussa T, Tuure T, Kuitunen M 2007 Probiotics and prebiotic galacto-oligosaccharides in the prevention of allergic diseases: a randomized, double-blind, placebo-controlled trial. J Allergy Clin Immunol 119: 192–198
Marzotto M, Maffeis C, Paternoster T, Ferrario R, Rizzotti L, Pellegrino M, Dellaglio F, Torriani S 2006 Lactobacillus paracasei A survives gastrointestinal passage and affects the fecal microbiota of healthy infants. Res Microbiol 157: 857–866
Atlas RM 1999 Probiotics—snake oil for the new millennium?. Environ Microbiol 1: 377–382
Tannock GW, Munro K, Harmsen HJ, Welling GW, Smart Gopal JK 2000 Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl Environ Microbiol 66: 2578–2588
Rawls JF, Mahowald MA, Ley RE, Gordon JI 2006 Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127: 423–433
Acknowledgements
The authors thank Karen Munro and Or Ming Yan for technical assistance, and Dr Yvonne Ng Peng, Judy Anthony, Corinne Kwek Poh Lian, and Soh Shu E for their excellent work in the clinic. We also thank Nestle for providing us the infant formula with and without probiotic bacteria, and preparing the randomization list. The voluntary participation of all subjects in the study is sincerely appreciated.
Author information
Authors and Affiliations
Additional information
The study was supported by National Medical Research Council (NMRC0971/2005), Republic of Singapore.
Rights and permissions
About this article
Cite this article
Mah, K., Chin, V., Wong, W. et al. Effect of a Milk Formula Containing Probiotics on the Fecal Microbiota of Asian Infants at Risk of Atopic Diseases. Pediatr Res 62, 674–679 (2007). https://doi.org/10.1203/PDR.0b013e31815991d5
Received:
Accepted:
Issue date:
DOI: https://doi.org/10.1203/PDR.0b013e31815991d5
This article is cited by
-
Compositional Quality and Possible Gastrointestinal Performance of Marketed Probiotic Supplements
Probiotics and Antimicrobial Proteins (2022)
-
The Role of Probiotics in the Prevention and Treatment of Atopic Dermatitis in Children: An Updated Systematic Review and Meta-Analysis of Randomized Controlled Trials
Pediatric Drugs (2020)
-
Bifidobacterium longum subsp infantis CECT7210-supplemented formula reduces diarrhea in healthy infants: a randomized controlled trial
Pediatric Research (2018)
-
Molecular analysis of infant fecal microbiota in an Asian at-risk cohort–correlates with infant and childhood eczema
BMC Research Notes (2014)
-
Association between funding source, methodological quality and research outcomes in randomized controlled trials of synbiotics, probiotics and prebiotics added to infant formula: A Systematic Review
BMC Medical Research Methodology (2013)