Abstract
Background:
Intrauterine growth restriction (IUGR) is a frequent complication of pregnancy defined as a restriction of fetal growth. The objective of this work was to improve the knowledge on the pathophysiology of IUGR using a genome-wide method of expression analysis.
Methods:
We analyzed differentially expressed genes in pooled placental tissues from vascular IUGR (four pools of three placentas) and normal pregnancies (four pools of three placentas) using a long nucleotide microarray platform (Nimblegen). We first did a global bioinformatics analysis based only on P value without any a priori. We secondly focused on “target” genes among the most modified ones. Finally, reverse transcription quantitative polymerase chain reaction (RT-qPCR) was performed on an extended panel of tissue samples (n = 62) on selected “target”.
Results:
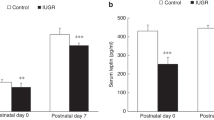
We identified 636 modified genes among which 206 were upregulated (1.5 and higher; P < 0.05). Groups of patients were classified unambiguously. Genes involved in mitochondrial function and oxidative phosphorylation were decreased affecting three out of five complexes of the respiratory chain of the mitochondria, and thus energy production and metabolism. Among the most induced genes, we identified LEP, IGFBP1, and RBP4.
Conclusion:
Complementary studies on the role and function of LEP, IGFBP1, and RBP4 in IUGR pathophysiology and also in fetal programming remain necessary.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Charkaluk ML, Marchand-Martin L, Ego A, et al.; Epipage Study Group. The influence of fetal growth reference standards on assessment of cognitive and academic outcomes of very preterm children. J Pediatr 2012;161:1053–8.
American College of Obstetricians and Gynecologists. ACOG Practice bulletin no. 134: fetal growth restriction. Obstet Gynecol 2013;121:1122–33.
Zeitlin J, El Ayoubi M, Jarreau PH, et al.; MOSAIC Research Group. Impact of fetal growth restriction on mortality and morbidity in a very preterm birth cohort. J Pediatr 2010;157:733–9.e1.
McIntire DD, Bloom SL, Casey BM, Leveno KJ. Birth weight in relation to morbidity and mortality among newborn infants. N Engl J Med 1999;340:1234–8.
Damodaram M, Story L, Kulinskaya E, Rutherford M, Kumar S. Early adverse perinatal complications in preterm growth-restricted fetuses. Aust N Z J Obstet Gynaecol 2011;51:204–9.
Flamant C, Gascoin G. [Short-term outcome and small for gestational age newborn management]. J Gynecol Obstet Biol Reprod (Paris) 2013;42:985–95.
Myatt L. Placental adaptive responses and fetal programming. J Physiol 2006;572(Pt 1):25–30.
Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ. Weight in infancy and death from ischaemic heart disease. Lancet 1989;2:577–80.
Gascoin G, Flamant C. [Long-term outcome in context of intra uterine growth restriction and/or small for gestational age newborns]. J Gynecol Obstet Biol Reprod (Paris) 2013;42:911–20.
Buffat C, Mondon F, Rigourd V, et al. A hierarchical analysis of transcriptome alterations in intrauterine growth restriction (IUGR) reveals common pathophysiological pathways in mammals. J Pathol 2007;213:337–46.
Vaiman D, Calicchio R, Miralles F. Landscape of transcriptional deregulations in the preeclamptic placenta. PLoS One 2013;8:e65498.
Moslehi R, Mills JL, Signore C, Kumar A, Ambroggio X, Dzutsev A. Integrative transcriptome analysis reveals dysregulation of canonical cancer molecular pathways in placenta leading to preeclampsia. Sci Rep 2013;3:2407.
Winn VD, Haimov-Kochman R, Paquet AC, et al. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology 2007;148:1059–79.
Nishizawa H, Ota S, Suzuki M, et al. Comparative gene expression profiling of placentas from patients with severe pre-eclampsia and unexplained fetal growth restriction. Reprod Biol Endocrinol 2011;9:107.
Struwe E, Berzl G, Schild R, et al. Microarray analysis of placental tissue in intrauterine growth restriction. Clin Endocrinol (Oxf) 2010;72:241–7.
Kim J, Zhao K, Jiang P, et al. Transcriptome landscape of the human placenta. BMC Genomics 2012;13:115.
Shi Z, Long W, Zhao C, Guo X, Shen R, Ding H. Comparative proteomics analysis suggests that placental mitochondria are involved in the development of pre-eclampsia. PLoS One 2013;8:e64351.
Mandò C, De Palma C, Stampalija T, et al. Placental mitochondrial content and function in intrauterine growth restriction and preeclampsia. Am J Physiol Endocrinol Metab 2014;306:E404–13.
Edery P, Marcaillou C, Sahbatou M, et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science 2011;332:240–3.
Bajoria R, Sooranna SR, Ward BS, Chatterjee R. Prospective function of placental leptin at maternal-fetal interface. Placenta 2002;23:103–15.
Gambino YP, Maymó JL, Pérez Pérez A, Calvo JC, Sánchez-Margalet V, Varone CL. Elsevier Trophoblast Research Award lecture: Molecular mechanisms underlying estrogen functions in trophoblastic cells–focus on leptin expression. Placenta 2012;33 Suppl:S63–70.
Maymó JL, Pérez Pérez A, Gambino Y, Calvo JC, Sánchez-Margalet V, Varone CL. Review: Leptin gene expression in the placenta–regulation of a key hormone in trophoblast proliferation and survival. Placenta 2011;32:Suppl 2:S146–53.
Dötsch J, Nüsken KD, Knerr I, Kirschbaum M, Repp R, Rascher W. Leptin and neuropeptide Y gene expression in human placenta: ontogeny and evidence for similarities to hypothalamic regulation. J Clin Endocrinol Metab 1999;84:2755–8.
McCarthy JF, Misra DN, Roberts JM. Maternal plasma leptin is increased in preeclampsia and positively correlates with fetal cord concentration. Am J Obstet Gynecol 1999;180(3 Pt 1):731–6.
Mise H, Yura S, Itoh H, et al. The relationship between maternal plasma leptin levels and fetal growth restriction. Endocr J 2007;54:945–51.
Breier BH, Vickers MH, Ikenasio BA, Chan KY, Wong WP. Fetal programming of appetite and obesity. Mol Cell Endocrinol 2001;185:73–9.
Lee PD, Giudice LC, Conover CA, Powell DR. Insulin-like growth factor binding protein-1: recent findings and new directions. Proc Soc Exp Biol Med 1997;216:319–57.
Leu JI, Crissey MA, Taub R. Massive hepatic apoptosis associated with TGF-beta1 activation after Fas ligand treatment of IGF binding protein-1-deficient mice. J Clin Invest 2003;111:129–39.
Reinehr T, Kleber M, Toschke AM, Woelfle J, Roth CL. Longitudinal association between IGFBP-1 levels and parameters of the metabolic syndrome in obese children before and after weight loss. Int J Pediatr Obes 2011;6:236–43.
Chan TF, Tsai YC, Wu CH, Lee CH, Wang SH, Su JH. The positive correlation between cord serum retinol-binding protein 4 concentrations and fetal growth. Gynecol Obstet Invest 2011;72:98–102.
Miles JL, Huber K, Thompson NM, Davison M, Breier BH. Moderate daily exercise activates metabolic flexibility to prevent prenatally induced obesity. Endocrinology 2009;150:179–86.
Meisinger C, Rückert IM, Rathmann W, et al. Retinol-binding protein 4 is associated with prediabetes in adults from the general population: the Cooperative Health Research in the Region of Augsburg (KORA) F4 Study. Diabetes Care 2011;34:1648–50.
Rhie YJ, Choi BM, Eun SH, Son CS, Park SH, Lee KH. Association of serum retinol binding protein 4 with adiposity and pubertal development in Korean children and adolescents. J Korean Med Sci 2011;26:797–802.
Mamelle N, Boniol M, Rivière O, et al. Identification of newborns with Fetal Growth Restriction (FGR) in weight and/or length based on constitutional growth potential. Eur J Pediatr 2006;165:717–25.
Gascoin-Lachambre G, Buffat C, Rebourcet R, et al. Cullins in human intra-uterine growth restriction: expressional and epigenetic alterations. Placenta 2010;31:151–7.
Barbaux S, Gascoin-Lachambre G, Buffat C, et al. A genome-wide approach reveals novel imprinted genes expressed in the human placenta. Epigenetics 2012;7:1079–90.
Vaiman D, Gascoin-Lachambre G, Boubred F, et al. The intensity of IUGR-induced transcriptome deregulations is inversely correlated with the onset of organ function in a rat model. PLoS One 2011;6:e21222.
Meller M, Vadachkoria S, Luthy DA, Williams MA. Evaluation of housekeeping genes in placental comparative expression studies. Placenta 2005;26:601–7.
Glaab E, Garibaldi JM, Krasnogor N. ArrayMining: a modular web-application for microarray analysis combining ensemble and consensus methods with cross-study normalization. BMC Bioinformatics 2009;10:358.
Acknowledgements
We are grateful to all participating patients, as well as to the Centre d’Investigation Clinique en Périnatalogie of the Port Royal maternity for their expertise and help.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Figure S1
(TIFF 1609 kb)
Supplementary Tables
(DOC 694 kb)
Rights and permissions
About this article
Cite this article
Madeleneau, D., Buffat, C., Mondon, F. et al. Transcriptomic analysis of human placenta in intrauterine growth restriction. Pediatr Res 77, 799–807 (2015). https://doi.org/10.1038/pr.2015.40
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/pr.2015.40
This article is cited by
-
Obesity downregulates lipid metabolism genes in first trimester placenta
Scientific Reports (2022)
-
Metformin and insulin treatment of gestational diabetes: effects on inflammatory markers and IGF-binding protein-1 – secondary analysis of a randomized controlled trial
BMC Pregnancy and Childbirth (2020)
-
DLX3 interacts with GCM1 and inhibits its transactivation-stimulating activity in a homeodomain-dependent manner in human trophoblast-derived cells
Scientific Reports (2017)
-
Sildenafil Therapy Normalizes the Aberrant Metabolomic Profile in the Comt−/− Mouse Model of Preeclampsia/Fetal Growth Restriction
Scientific Reports (2015)