Abstract
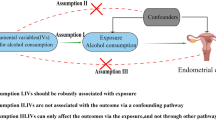
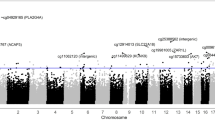
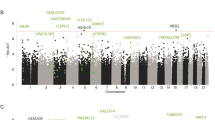
Epigenetic factors and consumption of alcohol, which suppresses DNA methylation, may influence the development and progression of epithelial ovarian cancer (EOC). However, there is a lack of understanding whether these factors interact to affect the EOC risk. In this study, we aimed to gain insight into this relationship by identifying leukocyte-derived DNA methylation markers acting as potential mediators of alcohol-associated EOC. We implemented a causal inference test (CIT) and the VanderWeele and Vansteelandt multiple mediator model to examine CpG sites that mediate the association between alcohol consumption and EOC risk. We modified one step of the CIT by adopting a high-dimensional inference procedure. The data were based on 196 cases and 202 age-matched controls from the Mayo Clinic Ovarian Cancer Case-Control Study. Implementation of the CIT test revealed two CpG sites (cg09358725, cg11016563), which represent potential mediators of the relationship between alcohol consumption and EOC case–control status. Implementation of the VanderWeele and Vansteelandt multiple mediator model further revealed that these two CpGs were the key mediators. Decreased methylation at both CpGs was more common in cases who drank alcohol at the time of enrollment vs. those who did not. cg11016563 resides in TRPC6 which has been previously shown to be overexpressed in EOC. These findings suggest two CpGs may serve as novel biomarkers for EOC susceptibility.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Morgan RJ Jr., et al. Epithelial ovarian cancer. J Natl Compr Cancer Netw. 2011;9:82–113.
Bagnardi V, Blangiardo M, La Vecchia C, Corrao G. A meta-analysis of alcohol drinking and cancer risk. Br J Cancer. 2001;85:1700–5.
Allen N, Anderson LM, Beland FA, Bénichou J, Beral V, Bloomfield K, et al. Alcohol consumption and ethyl carbamate. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. 2010;96:1–1379.
Bagnardi V, et al. Alcohol consumption and site-specific cancer risk: a comprehensive dose–response meta-analysis. Br J Cancer. 2015;112:580–93.
Yan-Hong H, et al. Association between alcohol consumption and the risk of ovarian cancer: a meta-analysis of prospective observational studies. BMC Public Health. 2015;15:223.
Rota M, et al. Alcohol drinking and epithelial ovarian cancer risk. A systematic review and meta-analysis. Gynecol Oncol. 2012;125:758–63.
Kelemen LE, et al. Recent alcohol consumption and risk of incident ovarian carcinoma: a pooled analysis of 5,342 cases and 10,358 controls from the Ovarian Cancer Association Consortium. BMC Cancer. 2013;13:28.
Genkinger J, et al. Alcohol intake and ovarian cancer risk: a pooled analysis of 10 cohort studies. Br J Cancer. 2006;94:757–62.
Cook LS, et al. Adult lifetime alcohol consumption and invasive epithelial ovarian cancer risk in a population-based case–control study. Gynecol Oncol. 2016;140:277–84.
Webb PM, Purdie DM, Bain CJ, Green AC. Alcohol, wine, and risk of epithelial ovarian cancer. Cancer Epidemiol Prev Biomarkers. 2004;13:592–9.
Ramchandani S, Bhattacharya SK, Cervoni N, Szyf M. DNA methylation is a reversible biological signal. Proc Natl Acad Sci. 1999;96:6107–12.
Cvetkovic D. Early events in ovarian oncogenesis. Reprod Biol Endocrinol. 2003;1:68.
Umeora M. Epigenetics and fetal alcohol spectrum disorders (FASD). Ebonyi Med J. 2012;11:1–6.
Philibert R, Plume JM, Gibbons FX, Brody GH, Beach S. The impact of recent alcohol use on genome wide DNA methylation signatures. Front Genet. 2012;3:54.
Bauerschlag DO, et al. Progression-free survival in ovarian cancer is reflected in epigenetic DNA methylation profiles. Oncology. 2011;80:12–20.
Watts GS, et al. DNA methylation changes in ovarian cancer are cumulative with disease progression and identify tumor stage. BMC Med Genomics. 2008;1:47.
Shen H, et al. Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat Commun. 2013;4:1628.
Cortessis VK, et al. Environmental epigenetics: prospects for studying epigenetic mediation of exposure–response relationships. Hum Genet. 2012;131:1565–89.
Millstein J, Zhang B, Zhu J, Schadt EE. Disentangling molecular relationships with a causal inference test. BMC Genet. 2009;10:23.
Fan J, Han F, Liu H. Challenges of big data analysis. Natl Sci Rev. 2014;1:293–314.
Dezeure R, Bühlmann P, Meier L, Meinshausen N. High-dimensional inference: confidence intervals, p-values and R-software hdi. Stat. Sci. 2015;30:533–58.
Koestler DC, et al. Integrative genomic analysis identifies epigenetic marks that mediate genetic risk for epithelial ovarian cancer. BMC Med Genomics. 2014;7:8.
Goode EL, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010;42:874–9.
Du P, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587.
Houseman EA, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86.
MacKinnon DP. Introduction to statistical mediation analysis. New York: Lawrence Erlbaum Associates; 2008.
Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Series B (Methodol). 1996;58:267–88.
Van de Geer S, Bühlmann P, Ritov Ya, Dezeure R. On asymptotically optimal confidence regions and tests for high-dimensional models. Ann Stat. 2014;42:1166–202.
VanderWeele TJ, Vansteelandt S. Mediation analysis with multiple mediators. Epidemiol Methods. 2014;2:95–115.
Varela-Rey M, Woodhoo A, Martinez-Chantar M-L, Mato JM, Lu SC. Alcohol, DNA methylation, and cancer. Alcohol Res. 2013;35:25–35.
Ungerer M, Knezovich J, Ramsay M. In utero alcohol exposure, epigenetic changes, and their consequences. Alcohol Res. 2013;35:37–46.
Shukla SD, Lim RW. Epigenetic effects of ethanol on the liver and gastrointestinal system. Alcohol Res. 2013;35:47–55.
Schernhammer ES, et al. Dietary folate, alcohol and B vitamins in relation to LINE-1 hypomethylation in colon cancer. Gut. 2010;59:794–9.
Christensen BC, et al. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet. 2010;6:e1001043.
Yu Y, et al. Enhanced expression of transient receptor potential channels in idiopathic pulmonary arterial hypertension. Proc Natl Acad Sci USA. 2004;101:13861–6.
Thébault S, et al. Receptor‐operated Ca2+ entry mediated by TRPC3/TRPC6 proteins in rat prostate smooth muscle (PS1) cell line. J Cell Physiol. 2005;204:320–8.
Zeng B, Yuan C, Yang X, L Atkin S, Xu S-Z. TRPC channels and their splice variants are essential for promoting human ovarian cancer cell proliferation and tumorigenesis. Curr Cancer Drug Targets. 2013;13:103–16.
Lossos IS, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–37.
Ma S, et al. The significance of LMO2 expression in the progression of prostate cancer. J Pathol. 2007;211:278–85.
Nakata K, et al. LMO2 is a novel predictive marker for a better prognosis in pancreatic cancer. Neoplasia. 2009;11:712–9.
Pearce N. Analysis of matched case-control studies. BMJ. 2016;352:i969.
Sun T, Zhang C-H. Scaled sparse linear regression. Biometrika. 2012;99:879–898.
Acknowledgements
We wish to thank an associate editor and two anonymous referees for their insightful comments that greatly improved the manuscript. This work was supported in part by grants from NSFC (31371336) and NIH (R01-CA122443).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Wu, D., Yang, H., Winham, S.J. et al. Mediation analysis of alcohol consumption, DNA methylation, and epithelial ovarian cancer. J Hum Genet 63, 339–348 (2018). https://doi.org/10.1038/s10038-017-0385-8
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s10038-017-0385-8