Abstract
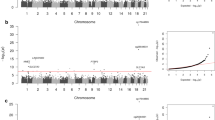
Diabetes-related hearing loss (DRHL) is a complication of diabetes mellitus that is drawing more attention currently. DNA methylation has a critical role in the pathogenesis of type 2 diabetes mellitus (T2DM) and its complications. Therefore, we investigated the genome-wide DNA methylation of peripheral blood of T2DM patients with/without hearing loss in order to explore the susceptibility loci of DRHL. Between DRHL group and control group, 113 gene sites were identified to be differentially methylated regions (DMRs). Among 38 DMRs with whole samples, the classification accuracy is up to 90%. With alignment to T2DM susceptibility genes and deafness genes published, KCNJ11 was found to be the only overlapped gene. The DNA methylation level of KCNJ11 was associated with stroke (t = 2.595, p < 0.05), but not with diabetic nephropathy and diabetic retinopathy. The detective rate of distortion product otoacoustic emissions (DPOAE) from low to high frequencies (0.7–6 kHz) on the right ear was significantly correlated with the methylation level of KCNJ11. The auditory brainstem response (ABR) threshold on the right ear was also correlated (r = 0.678, p < 0.05). This DNA methylation profile indicates the susceptibility loci of DRHL. The potassium metabolism may have a critical role in the hearing loss caused by hyperglycemia.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bainbridge KE, Hoffman HJ, Cowie CC. Diabetes and hearing impairment in the United States: audiometric evidence from the National Health and Nutrition Examination Survey, 1999 to 2004. Ann Intern Med. 2008 ;149:1–10.
Cheng YJ, Gregg EW, Saaddine JB, Imperatore G, Zhang X, Albright AL. Three decade change in the prevalence of hearing impairment and its association with diabetes in the United States. Prev Med. 2009;49:360–4.
Sakuta H, Suzuki T, Yasuda H, Ito T. Type 2 diabetes and hearing loss in personnel of the self-defense forces. Diabetes Res Clin Pract. 2007;75:229–34.
Wang L, Gao P, Zhang M, Huang Z, Zhang D, Deng Q, et al. Prevalence and ethnic pattern of diabetes and prediabetes in China in 2013. JAMA. 2017;317:2515–23.
Xu GC, Luo Y, Li Q, Wu MF, Zhou ZJ. Standardization of type 2 diabetes outpatient expenditure with bundled payment method in China. Chin Med J. 2016;129:953–9.
Chen XB, Tang L, Chen HY, Zhao LY, Hu SL. Assessing the impact of complications on the costs of type 2 diabetes in urban China. Chinese. J Diabetes. 2003;11:238–41.
Hao J, Fu X, Zhang C, Zhang X, Zhao S, Li Y. Early detection of hearing impairment in patients with diabetes mellitus with otoacoustic emission. A systematic review and meta-analysis. Acta Otolaryngol. 2017;137:179–85.
Multhaup ML, Seldin MM, Jaffe AE, Lei X, Kirchner H, Mondal P, et al. Mouse-human experimental epigenetic analysis unmasks dietary targets and genetic liability for diabetic phenotypes. Cell Metab. 2015;21:138–49.
Sapienza C, Lee J, Powell J, Erinle O, Yafai F, Reichert J, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2011;6:20–8.
Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci USA 2010;107(Suppl 1):1757–64.
Toperoff G, Aran D, Kark JD, Rosenberg M, Dubnikov T, Nissan B, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21:371–83.
American Diabetes Association. 2. Classification and diagnosis of diabetes. Diabetes Care. 2017;40(Suppl 1):S11–S24.
Dubno JR, Eckert MA, Lee FS, Matthews LJ, Schmiedt RA. Classifying human audiometric phenotypes of age-related hearing loss from animal models. J Assoc Res Otolaryngol. 2013;14:687–701.
Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–81.
Prokopenko I, McCarthy MI, Lindgren CM. Type 2 diabetes: new genes, new understanding. Trends Genet. 2008;24:613–21.
Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–89.
Bonnefond A, Froguel P, Vaxillaire M. The emerging genetics of type 2 diabetes. Trends Mol Med. 2010;16:407–16.
Ridderstråle M, Groop L. Genetic dissection of type 2 diabetes. Mol Cell Endocrinol. 2009;297:10–7.
Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90.
Cho YS, Chen CH, Hu C, Long J, Ong RT, Sim X, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2011;44:67–72.
Hara K, Fujita H, Johnson TA, Yamauchi T, Yasuda K, Horikoshi M, et al. Genome-wide association study identifies three novel loci for type 2 diabetes. Hum Mol Genet. 2014;23:239–46.
DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; Mexican American Type 2 Diabetes (MAT2D) Consortium; Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium, Mahajan A, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–44.
Ayub Q, Moutsianas L, Chen Y, Panoutsopoulou K, Colonna V, Pagani L, et al. Revisiting the thrifty gene hypothesis via 65 loci associated with susceptibility to type 2 diabetes. Am J Hum Genet. 2014;94:176–85.
Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38.
Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600.
Mutai H, Miya F, Fujii M, Tsunoda T, Matsunaga T. Attenuation of progressive hearing loss in DBA/2J mice by reagents that affect epigenetic modifications is associated with up-regulation of the zinc importer Zip4. PLoS ONE. 2015;10:e0124301.
Olson TM, Terzic A. Human K(ATP) channelopathies: diseases of metabolic homeostasis. Pflug Arch. 2010;460:295–306.
Bonnefond A, Philippe J, Durand E, Dechaume A, Huyvaert M, Montagne L, et al. Whole-exome sequencing and high throughput genotyping identified KCNJ11 as the thirteenth MODY gene. PLoS ONE. 2012;7:e37423.
Gabashvili IS, Sokolowski BH, Morton CC, Giersch AB. Ion channel gene expression in the inner ear. J Assoc Res Otolaryngol. 2007;8:305–28.
Chen J, Zhao HB. The role of an inwardly rectifying K(+) channel (Kir4.1) in the inner ear and hearing loss. Neuroscience. 2014;265:137–46.
Jacobs PG, Konrad-Martin D, McMillan GP, McDermott D, Fausti SA, Kagen D, et al. Influence of acute hyperglycemia on otoacoustic emissions and the medial olivocochlear reflex. J Acoust Soc Am. 2012;131:1296–306.
Wolber LE, Steves CJ, Tsai PC, Deloukas P, Spector TD, Bell JT, et al. Epigenome-wide DNA methylation in hearing ability: new mechanisms for an old problem. PLoS ONE. 2014;9:e105729.
Martin GM, Yoshioka C, Rex EA, Fay JF, Xie Q, Whorton MR, et al. Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating. Elife. 2017;6:e24149.
Acknowledgements
We appreciate the support of all the patients and their relatives in our study. Without their understanding and generous contribution, this study cannot be implemented. We also thank HXJ for screening charts in the electrical chart system, which is a tremendous working load. We are grateful that HXL, WMX, CHL and their colleagues have undertaken a great amount of lab work (Genesky Biotechnologies Inc., Shanghai, China, 201315). This work was supported by Basic-Clinical Cooperation Program from Capital Medical University (No. 16JL58) for Dr. Jin Hao.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Co-corresponding author: Yongxin Li
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Hao, J., Hua, L., Fu, X. et al. Genome-wide DNA methylation analysis of human peripheral blood reveals susceptibility loci of diabetes-related hearing loss. J Hum Genet 63, 1241–1250 (2018). https://doi.org/10.1038/s10038-018-0507-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-018-0507-y
This article is cited by
-
A systematic review on the contribution of DNA methylation to hearing loss
Clinical Epigenetics (2024)
-
Diabetes-induced auditory complications: are they preventable? a comprehensive review of interventions
European Archives of Oto-Rhino-Laryngology (2021)