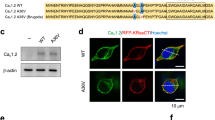
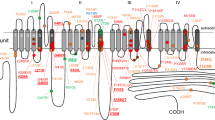
Abstract
Sinoatrial node dysfunction and deafness (SANDD) syndrome is rare and characterized by a low heart beat and severe-to-profound deafness. Additional features include fatigue, dizziness, and episodic syncope. The sinoatrial node (SAN) drives heart automaticity and continuously regulates heart rate. The CACNA1D gene encoding the Cav1.3 protein expressed in inner hair cells, atria and SAN, induces loss-of-function in channel activity and underlies SANDD. To date, only one variant c.1208_1209insGGG:p.(G403_V404insG) has been reported for SANDD syndrome. We studied five Pakistani families with SANDD and characterized a new missense variant p.(A376V) in CACNA1D in one family, and further characterized the founder variant p.(G403_V404insG) in four additional pedigrees. We show that affected individuals in the four families which segregate p.(G403_V404insG) share a 1.03 MB haplotype on 3p21.1 suggesting they share a common distant ancestor. In conclusion, we identified new and known variants in CACNA1D in five Pakistani families with SANDD. This study is of clinical importance as the CACNA1D founder variant is only observed in families from the Khyber Pakhtunkhwa (KPK) province, in Pakistan. Therefore, screening patients with congenital deafness for SAN dysfunction in this province could ensure adequate follow-up and prevent cardiac failure associated with SAN.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Morton CC, Nance WE. Newborn hearing screening—a silent revolution. N Engl J Med. 2006;354:2151–64.
Baig SM, Koschak A, Lieb A, Gebhart M, Dafinger C, Nürnberg G, et al. Loss of Ca(v)1.3 (CACNA1D) function in a human channelopathy with bradycardia and congenital deafness. Nat Neurosci. 2011;14:77–84.
Platzer J, Engel J, Schrott-Fischer A, Stephan K, Bova S, Chen H, et al. Congenital deafness and sinoatrial node dysfunction in mice lacking class D L-type Ca2+ channels. Cell. 2000;102:89–97.
Marger L, Mesirca P, Alig J, Torrente A, Dübel S, Engeland B, et al. Functional roles of Ca v 1.3, Ca v 3.1 and HCN channels in automaticity of mouse atrioventricular cells: insights into the atrioventricular pacemaker mechanism. Channels. 2011;5:251–61.
Catterall WA. Signaling complexes of voltage-gated sodium and calcium channels. Neurosci Lett. 2010;486:107–16.
Benson DW, Wang DW, Dyment M, Knilans TK, Fish FA, Strieper MJ, et al. Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A). J Clin Invest. 2003;112:1019–28.
Satheesh SV, Kunert K, Ruttiger L, Zuccotti A, Schonig K, Friauf E, et al. Retrocochlear function of the peripheral deafness gene Cacna1d. Hum Mol Genet. 2012;21:3896–909.
Green MR, Sambrook J. Isolation of high-molecular-weight DNA using organic solvents. Cold Spring Harb Protoc. 2017; 2017:356–9.
Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101.
Seelow D, Schuelke M. HomozygosityMapper2012—bridging the gap between homozygosity mapping and deep sequencing. Nucleic Acids Res. 2012;40:W516–20.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–95.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164.
Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–9.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–91.
Chen C, Huang H, Wu CH. Protein bioinformatics databases and resources. Methods Mol Biol. 2017;1558:3–39.
The PyMOL Molecular Graphics System. Version 2.0. Schrödinger, LLC, New York, NY, USA. 2017.
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera? A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12.
Greater Middle East Variome Consortium, Scott EM, Halees A, Itan Y, Spencer EG, He Y, et al. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet. 2016;48:1071–6.
Exome Aggregation Consortium, Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Striessnig J, Pinggera A, Kaur G, Bock G, Tuluc P. L-type Ca2+ channels in heart and brain. Wiley Interdiscip Rev Membr Transp Signal. 2014;3:15–38.
Dirksen RT, Nakai J, Gonzalez A, Imoto K, Beam KG. The S5-S6 linker of repeat I is a critical determinant of L-type Ca2 + channel conductance. Biophys J. 1997;73:1402–9.
Mangoni ME, Couette B, Bourinet E, Platzer J, Reimer D, Striessnig J, et al. Functional role of L-type Cav1.3 Ca2+ channels in cardiac pacemaker activity. Proc Natl Acad Sci USA. 2003;100:5543–8.
Cavalli-Sforza LL, Feldman MW. The application of molecular genetic approaches to the study of human evolution. Nat Genet. 2003;33(Suppl):266–75.
Acknowledgements
We would like to thank the families for their participation in this study. This work was supported by the Higher Education Commission of Pakistan (to W.A.) and National Institutes of Health (NIH)-National Institute of Deafness and other Disorders grants R01 DC011651 and R01 DC003594 (to S.M.L). “Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268201200008I”. Exome sequencing performed at the University of Washington Center for Mendelian Genomics was funded by the NIH–National Human Genome Research Institute grant UM1 HG006493 (to D.A.N., M.J.B., and S.M.L.).
Supplementary data
The Supplementary information in this paper includes the list of members of the University of Washington Center for Mendelian Genomics (UWCMG) study group and two figures (Supplementary figure 1; Supplementary figure 2).
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
10038_2018_542_MOESM1_ESM.docx
Supplementary data: The Supplementary information in this paper includes the list of members of the University of Washington Center for Mendelian Genomics (UWCMG) study group and two figures (Supplementary figure 1; Supplementary figure 2).
Rights and permissions
About this article
Cite this article
Liaqat, K., Schrauwen, I., Raza, S.I. et al. Identification of CACNA1D variants associated with sinoatrial node dysfunction and deafness in additional Pakistani families reveals a clinical significance. J Hum Genet 64, 153–160 (2019). https://doi.org/10.1038/s10038-018-0542-8
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s10038-018-0542-8
This article is cited by
-
A Systematic Review on the Role of the Stria Vascularis in Menière’s Disease Pathogenesis
Journal of the Association for Research in Otolaryngology (2025)
-
Pathogenicity of de novo CACNA1D Ca2+ channel variants predicted from sequence co-variation
European Journal of Human Genetics (2024)
-
Fine mapping of candidate effector genes for heart rate
Human Genetics (2024)
-
Spatially resolved multiomics of human cardiac niches
Nature (2023)
-
Molecular genetic landscape of hereditary hearing loss in Pakistan
Human Genetics (2022)