Abstract
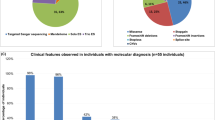
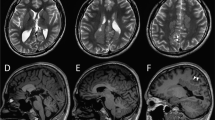
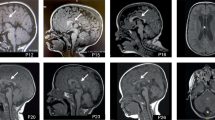
Casein kinase 2 (CK2) is a serine threonine kinase ubiquitously expressed in eukaryotic cells and involved in various cellular processes. In recent studies, de novo variants in CSNK2A1 and CSNK2B, which encode the subunits of CK2, have been identified in individuals with intellectual disability syndrome. In this study, we describe four patients with neurodevelopmental disorders possessing de novo variants in CSNK2A1 or CSNK2B. Using whole-exome sequencing, we detected two de novo variants in CSNK2A1 in two unrelated Japanese patients, a novel variant c.571C>T, p.(Arg191*) and a recurrent variant c.593A>G, p.(Lys198Arg), and two novel de novo variants in CSNK2B in Japanese and Malaysian patients, c.494A>G, p.(His165Arg) and c.533_534insGT, p.(Pro179Tyrfs*49), respectively. All four patients showed mild to profound intellectual disabilities, developmental delays, and various types of seizures. This and previous studies have found a total of 20 CSNK2A1 variants in 28 individuals with syndromic intellectual disability. The hotspot variant c.593A>G, p.(Lys198Arg) was found in eight of 28 patients. Meanwhile, only five CSNK2B variants were identified in five individuals with neurodevelopmental disorders. We reviewed the previous literature to verify the phenotypic spectrum of CSNK2A1- and CSNK2B-related syndromes.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Harris JC. New classification for neurodevelopmental disorders in DSM-5. Curr Opin Psychiatry. 2014;27:95–97.
Soden SE, Saunders CJ, Willig LK, Farrow EG, Smith LD, Petrikin JE, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6:265ra168.
Miller AR, Masse LC, Shen J, Schiariti V, Roxborough L. Diagnostic status, functional status and complexity among Canadian children with neurodevelopmental disorders and disabilities: a population-based study. Disabil Rehabil. 2013;35:468–78.
Blencowe H, Lee AC, Cousens S, Bahalim A, Narwal R, Zhong N, et al. Preterm birth-associated neurodevelopmental impairment estimates at regional and global levels for 2010. Pediatr Res. 2013;74(Suppl 1):17–34.
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–8.
Lelieveld SH, Reijnders MR, Pfundt R, Yntema HG, Kamsteeg EJ, de Vries P, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19:1194–6.
de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9.
Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–7.
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21.
St-Denis NA, Litchfield DW. Protein kinase CK2 in health and disease: from birth to death: the role of protein kinase CK2 in the regulation of cell proliferation and survival. Cell Mol Life Sci. 2009;66:1817–29.
Guerra B, Issinger OG. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis. 1999;20:391–408.
Loizou JI, El-Khamisy SF, Zlatanou A, Moore DJ, Chan DW, Qin J, et al. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell. 2004;117:17–28.
Gotz C, Montenarh M. Protein kinase CK2 in development and differentiation. Biomed Rep. 2017;6:127–33.
Ghavidel A, Schultz MC. TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell. 2001;106:575–84.
Seldin DC, Landesman-Bollag E, Farago M, Currier N, Lou D, Dominguez I. CK2 as a positive regulator of Wnt signalling and tumourigenesis. Mol Cell Biochem. 2005;274:63–67.
Trembley JH, Wang G, Unger G, Slaton J, Ahmed K. Protein kinase CK2 in health and disease: CK2: a key player in cancer biology. Cell Mol Life Sci. 2009;66:1858–67.
Ahmad KA, Wang G, Unger G, Slaton J, Ahmed K. Protein kinase CK2--a key suppressor of apoptosis. Adv Enzym Regul. 2008;48:179–87.
Bodenbach L, Fauss J, Robitzki A, Krehan A, Lorenz P, Lozeman FJ, et al. Recombinant human casein kinase II. A study with the complete set of subunits (alpha, alpha’ and beta), site-directed autophosphorylation mutants and a bicistronically expressed holoenzyme. Eur J Biochem. 1994;220:263–73.
Duncan JS, Litchfield DW. Too much of a good thing: the role of protein kinase CK2 in tumorigenesis and prospects for therapeutic inhibition of CK2. Biochim Biophys Acta. 2008;1784:33–47.
Niefind K, Guerra B, Ermakowa I, Issinger OG. Crystal structure of human protein kinase CK2: insights into basic properties of the CK2 holoenzyme. EMBO J. 2001;20:5320–31.
Litchfield DW. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J. 2003;369:1–15.
Pinna LA. Protein kinase CK2. Int J Biochem Cell Biol. 1997;29:551–4.
Okur V, Cho MT, Henderson L, Retterer K, Schneider M, Sattler S, et al. De novo mutations in CSNK2A1 are associated with neurodevelopmental abnormalities and dysmorphic features. Hum Genet. 2016;135:699–705.
Owen CI, Bowden R, Parker MJ, Patterson J, Patterson J, Price S et al. Extending the phenotype associated with the CSNK2A1-related Okur-Chung syndrome-A clinical study of 11 individuals. Am J Med Genet A. 2018;176:1108–14.
Trinh J, Huning I, Budler N, Hingst V, Lohmann K, Gillessen-Kaesbach G. A novel de novo mutation in CSNK2A1: reinforcing the link to neurodevelopmental abnormalities and dysmorphic features. J Hum Genet. 2017;62:1005–6.
Chiu ATG, Pei SLC, Mak CCY, Leung GKC, Yu MHC, Lee SL et al. Okur-Chung neurodevelopmental syndrome: eight additional cases with implications on phenotype and genotype expansion. Clin Genet. 2018;93:880–90.
Poirier K, Hubert L, Viot G, Rio M, Billuart P, Besmond C, et al. CSNK2B splice site mutations in patients cause intellectual disability with or without myoclonic epilepsy. Hum Mutat. 2017;38:932–41.
Sakaguchi Y, Uehara T, Suzuki H, Kosaki K, Takenouchi T. Truncating mutation in CSNK2B and myoclonic epilepsy. Hum Mutat. 2017;38:1611–2.
Akahira-Azuma M, Tsurusaki Y, Enomoto Y, Mitsui J, Kurosawa K. Refining the clinical phenotype of Okur-Chung neurodevelopmental syndrome. Hum Genome Var. 2018;5:18011.
Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K, et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet. 2013;45:445–9, 449e441.
Fukai R, Saitsu H, Tsurusaki Y, Sakai Y, Haginoya K, Takahashi K, et al. De novo KCNH1 mutations in four patients with syndromic developmental delay, hypotonia and seizures. J Hum Genet. 2016;61:381–7.
Hiraide T, Nakashima M, Yamoto K, Fukuda T, Kato M, Ikeda H, et al. De novo variants in SETD1B are associated with intellectual disability, epilepsy and autism. Hum Genet. 2018;137:95–104.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Nagasaki M, Yasuda J, Katsuoka F, Nariai N, Kojima K, Kawai Y, et al. Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat Commun. 2015;6:8018.
Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Jagadeesh KA, Wenger AM, Berger MJ, Guturu H, Stenson PD, Cooper DN, et al. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet. 2016;48:1581–6.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Chakarova CF, Papaioannou MG, Khanna H, Lopez I, Waseem N, Shah A, et al. Mutations in TOPORS cause autosomal dominant retinitis pigmentosa with perivascular retinal pigment epithelium atrophy. Am J Hum Genet. 2007;81:1098–103.
Leachman NT, Brellier F, Ferralli J, Chiquet-Ehrismann R, Tucker RP. ATAD2B is a phylogenetically conserved nuclear protein expressed during neuronal differentiation and tumorigenesis. Dev Growth Differ. 2010;52:747–55.
Dominguez I, Degano IR, Chea K, Cha J, Toselli P, Seldin DC. CK2alpha is essential for embryonic morphogenesis. Mol Cell Biochem. 2011;356:209–16.
Lou DY, Dominguez I, Toselli P, Landesman-Bollag E, O’Brien C, Seldin DC. The alpha catalytic subunit of protein kinase CK2 is required for mouse embryonic development. Mol Cell Biol. 2008;28:131–9.
Xu X, Toselli PA, Russell LD, Seldin DC. Globozoospermia in mice lacking the casein kinase II alpha’ catalytic subunit. Nat Genet. 1999;23:118–21.
Cheusova T, Khan MA, Schubert SW, Gavin AC, Buchou T, Jacob G, et al. Casein kinase 2-dependent serine phosphorylation of MuSK regulates acetylcholine receptor aggregation at the neuromuscular junction. Genes Dev. 2006;20:1800–16.
Herrmann D, Straubinger M, Hashemolhosseini S. Protein kinase CK2 interacts at the neuromuscular synapse with Rapsyn, Rac1, 14-3-3gamma, and Dok-7 proteins and phosphorylates the latter two. J Biol Chem. 2015;290:22370–84.
Eiber N, Simeone L, Hashemolhosseini S. Ablation of protein kinase CK2beta in skeletal muscle fibers interferes with their oxidative capacity. Pharmaceuticals. 2017;10:13.
Kuntamalla PP, Kunttas-Tatli E, Karandikar U, Bishop CP, Bidwai AP. Drosophila protein kinase CK2 is rendered temperature-sensitive by mutations of highly conserved residues flanking the activation segment. Mol Cell Biochem. 2009;323:49–60.
Cosmelli D, Antonelli M, Allende CC, Allende JE. An inactive mutant of the alpha subunit of protein kinase CK2 that traps the regulatory CK2beta subunit. FEBS Lett. 1997;410:391–6.
Kusk M, Ahmed R, Thomsen B, Bendixen C, Issinger OG, Boldyreff B. Interactions of protein kinase CK2beta subunit within the holoenzyme and with other proteins. Mol Cell Biochem. 1999;191:51–58.
Raaf J, Issinger OG, Niefind K. First inactive conformation of CK2 alpha, the catalytic subunit of protein kinase CK2. J Mol Biol. 2009;386:1212–21.
Blond O, Jensen HH, Buchou T, Cochet C, Issinger OG, Boldyreff B. Knocking out the regulatory beta subunit of protein kinase CK2 in mice: gene dosage effects in ES cells and embryos. Mol Cell Biochem. 2005;274:31–37.
Buchou T, Vernet M, Blond O, Jensen HH, Pointu H, Olsen BB, et al. Disruption of the regulatory beta subunit of protein kinase CK2 in mice leads to a cell-autonomous defect and early embryonic lethality. Mol Cell Biol. 2003;23:908–15.
Marin O, Meggio F, Sarno S, Pinna LA. Physical dissection of the structural elements responsible for regulatory properties and intersubunit interactions of protein kinase CK2 beta-subunit. Biochemistry. 1997;36:7192–8.
Acknowledgements
We would like to thank the patients’ families for participating in this work. This study was supported by grants for: Research on Measures for Intractable Diseases from Ministry of Health, Labour and Welfare of Japan; Comprehensive Research on Disability Health and Welfare, the Strategic Research Program for Brain Science; Initiative on Rare and Undiagnosed Diseases in Pediatrics from the Japan Agency for Medical Research and Development; Grant-in-Aid for Scientific Research (A)(17H01539), (B)(16H05160), and (C)(15K10367) from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Nakashima, M., Tohyama, J., Nakagawa, E. et al. Identification of de novo CSNK2A1 and CSNK2B variants in cases of global developmental delay with seizures. J Hum Genet 64, 313–322 (2019). https://doi.org/10.1038/s10038-018-0559-z
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-018-0559-z
This article is cited by
-
Casein kinase 2 maintains the self-renewal of neural stem cells via prospero phosphorylation in Drosophila
Cell & Bioscience (2026)
-
Genetic analysis of four cases of Poirier Bienvenu neurodevelopmental syndrome associated with CSNK2B variant
BMC Medical Genomics (2025)
-
Characterization of 13 Novel Genetic Variants in Genes Associated with Epilepsy: Implications for Targeted Therapeutic Strategies
Molecular Diagnosis & Therapy (2024)
-
Comprehensive reanalysis for CNVs in ES data from unsolved rare disease cases results in new diagnoses
npj Genomic Medicine (2024)
-
Protein kinase CK2 phosphorylates a conserved motif in the Notch effector E(spl)-Mγ
Molecular and Cellular Biochemistry (2023)