Abstract
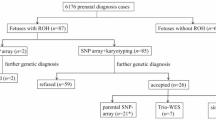
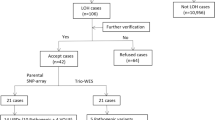
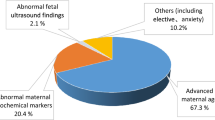
Region of homozygosity (ROH) is classified as uniparental disomy (UPD) or identity by descent, depending on its origin. To explore the clinical relevance of ROH in prenatal diagnoses, we reviewed 5063 fetal samples subjected to single nucleotide polymorphism array at our center over 5 years. ROH cases meeting our reporting threshold were further analyzed. ROHs were detected in 22 fetuses (0.43%, 22/5063), of which, 77.3% (17/22) showed a ROH on a single chromosome and 22.7% (5/22) showed multiple ROHs on different chromosomes. Among 5063 fetuses undergoing invasive prenatal diagnoses owing to various indications, five cases were identified as UPDs with a rate of ~1/1000. We observed clinically relevant UPDs in two cases related to Prader–Willi syndrome and transient neonatal diabetes mellitus. Of note, one case showed 50% mosaicism for trisomy 2 in amniotic fluid, whereas a complete UPD (2) was observed in umbilical cord blood. Trio whole-exome sequencing was performed for three cases. Clinically relevant variants were identified in two cases, one of which, NM_000302:c.2071_2072insCC (p.R693Qfs*122) in PLOD1 located in the ROH, may be related to Ehlers-Danlos syndrome, kyphoscoliotic type, 1. Overall, 72.7% (16/22) of the ROH carriers showed ultrasound abnormalities, of whom eight (50%, 8/16) had adverse perinatal outcomes. Our study demonstrates that the clinical relevance of ROHs should be examined regarding fetuses with ROHs occurring on imprinted chromosomes or those derived from consanguineous parents in prenatal diagnoses; imprinting disorders and/or autosomal recessive diseases attributed to ROHs should be considered during genetic counseling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Kearney HM, Kearney JB, Conlin LK. Diagnostic implications of excessive homozygosity detected by SNP-based microarrays: consanguinity, uniparental disomy, and recessive single-gene mutations. Clin Lab Med. 2011;31:595–613.
Del Gaudio D, Shinawi M, Astbury C, Tayeh MK, Deak KL, Raca G, ACMG Laboratory Quality Assurance Committee. Diagnostic testing for uniparental disomy: a points to consider statement from the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2020;22:1133–41.
Rehder CW, David KL, Hirsch B, Toriello HV, Wilson CM, Kearney HM. American College of Medical Genetics and Genomics: standards and guidelines for documenting suspected consanguinity as an incidental finding of genomic testing. Genet Med: Off J Am Coll Med Genet. 2013;15:150–2.
Niida Y, Ozaki M, Shimizu M, Ueno K, Tanaka T. Classification of Uniparental Isodisomy Patterns That Cause Autosomal Recessive Disorders: Proposed Mechanisms of Different Proportions and Parental Origin in Each Pattern. Cytogenetic Genome Res. 2018;154:137–46.
Hansen WF, Bernard LE, Langlois S, Rao KW, Chescheir NC, Aylsworth AS, et al. Maternal uniparental disomy of chromosome 2 and confined placental mosaicism for trisomy 2 in a fetus with intrauterine growth restriction, hypospadias, and oligohydramnios. Prenat Diagn. 1997;17:443–50.
Scheuvens R, Begemann M, Soellner L, Meschede D, Raabe-Meyer G, Elbracht M, et al. Maternal uniparental disomy of chromosome 16 [upd(16)mat]: clinical features are rather caused by (hidden) trisomy 16 mosaicism than by upd(16)mat itself. Clin Genet. 2017;92:45–51.
Spence JE, Perciaccante RG, Greig GM, Willard HF, Ledbetter DH, Hejtmancik JF, et al. Uniparental disomy as a mechanism for human genetic disease. Am J Hum Genet. 1988;42:217–26.
Vetro A, Goidin D, Lesende I, Limongelli I, Ranzani GN, Novara F, et al. Diagnostic application of a capture based NGS test for the concurrent detection of variants in sequence and copy number as well as LOH. Clin Genet. 2018;93:545–56.
Guo T, Tan ZP, Chen HM, Zheng DY, Liu L, Huang XG, et al. An effective combination of whole-exome sequencing and runs of homozygosity for the diagnosis of primary ciliary dyskinesia in consanguineous families. Sci Rep. 2017;7:7905.
Xiao B, Wang L, Liu H, Fan Y, Xu Y, Sun Y, et al. Uniparental isodisomy caused autosomal recessive diseases: NGS-based analysis allows the concurrent detection of homogenous variants and copy-neutral loss of heterozygosity. Mol Genet Genomic Med. 2019;7:e00945.
Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST, Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–5.
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22:245–57.
Schroeder C, Sturm M, Dufke A, Mau-Holzmann U, Eggermann T, Poths S, et al. UPDtool: a tool for detection of iso- and heterodisomy in parent-child trios using SNP microarrays. Bioinformatics. 2013;29:1562–4.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23.
McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–72.
Hoppman N, Rumilla K, Lauer E, Kearney H, Thorland E. Patterns of homozygosity in patients with uniparental disomy: detection rate and suggested reporting thresholds for SNP microarrays. Genet Med. 2018;20:1522–7.
Inoue T, Yagasaki H, Nishioka J, Nakamura A, Matsubara K, Narumi S, et al. Molecular and clinical analyses of two patients with UPD(16)mat detected by screening 94 patients with Silver-Russell syndrome phenotype of unknown aetiology. J Med Genet. 2019;56:413–8.
Kotzot D, Utermann G. Uniparental disomy (UPD) other than 15: phenotypes and bibliography updated. Am J Med Genet A. 2005;136:287–305.
Eggermann T. Prenatal Detection of Uniparental Disomies (UPD): Intended and Incidental Finding in the Era of Next Generation Genomics. Genes (Basel). 2020;11:1454.
Nakka P, Pattillo Smith S, O’Donnell-Luria AH, McManus KF. 23andMe Research Team, Mountain JL, et al. Characterization of Prevalence and Health Consequences of Uniparental Disomy in Four Million Individuals from the General Population. Am J Hum Genet. 2019;105:921–32.
Matsubara K, Murakami N, Nagai T, Ogata T. Maternal age effect on the development of Prader-Willi syndrome resulting from upd(15)mat through meiosis 1 errors. J Hum Genet. 2011;56:566–71.
Robinson WP, Christian SL, Kuchinka BD, Peñaherrera MS, Das S, Schuffenhauer S, et al. Somatic segregation errors predominantly contribute to the gain or loss of a paternal chromosome leading to uniparental disomy for chromosome 15. Clin Genet. 2000;57:349–58.
Bittles A. Consanguinity and its relevance to clinical genetics. Clin Genet. 2001;60:89–98.
Yingjun X, Zhiyang H, Linhua L, Fangming S, Linhuan H, Jinfeng T. Chromosomal uniparental disomy 16 and fetal intrauterine growth restriction. Eur J Obstet Gynecol Reprod Biol. 2017;211:1–7.
Kotzot D. Prenatal testing for uniparental disomy: indications and clinical relevance. Ultrasound Obstet Gynecol. 2008;31:100–5.
Wilkins-Haug L, Quade B, Morton CC. Confined placental mosaicism as a risk factor among newborns with fetal growth restriction. Prenat Diagn. 2006;26:428–32.
Acknowledgements
We deeply thank the patients and family members who participated in this study.
Funding
This work was supported by grant from Fujian Provincial Health Technology Project (grant no. 2020GGB018); Fujian Provincial Maternity and Child Health Hospital (grant no. YCXM 19–15); Fujian Provincial Natural Science Foundation (grant no. 2019J01509); and Joints Funds for the Innovation of Science and Technology, Fujian Province (grant no. 2020Y9159).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Liang, B., Yu, D., Zhao, W. et al. Prenatal diagnosis of fetuses with region of homozygosity detected by single nucleotide polymorphism array: a retrospective cohort study. J Hum Genet 67, 629–638 (2022). https://doi.org/10.1038/s10038-022-01062-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-022-01062-9
This article is cited by
-
A retrospective single center analysis of fetuses with region of homozygosity detected by single nucleotide polymorphism array
Scientific Reports (2025)
-
Prenatal ultrasound findings and clinical outcomes of uniparental disomy: a retrospective study
BMC Pregnancy and Childbirth (2024)
-
Genetic testing for fetal loss of heterozygosity using single nucleotide polymorphism array and whole-exome sequencing
Scientific Reports (2024)