Abstract
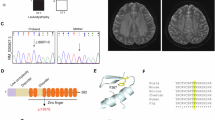
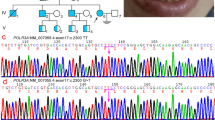
Hypomyelinating leukodystrophy-5 (HLD5) is a rare autosomal recessive hypomyelination disorder characterized by congenital cataract, progressive neurologic impairment, and myelin deficiency in the central and peripheral nervous system, caused by mutations in the HYCC1 gene. Here we report a 23-year-old girl with HLD5 from unrelated families. Molecular analysis was performed using sequence screening of the HYCC1 gene. In addition, in silico prediction tools and molecular investigation were used to predict the structural effect of the mutations. Results showed a novel compound heterozygous mutation in the HYCC1 gene. Moreover, in silico tools and 3D structural modeling revealed that c.521C > A (p.Ala174Glu) and c.652C > G (p.Gln218Glu) mutations could affect the structure, stability, and conformational analyses in the N-ter domain of the Hyccin protein. We also, we compared the phenotype of our patient with those of previously reported cases with HLD5 syndrome and our findings indicate the absence of reliable genotype-phenotype correlations. To the best of our knowledge, this is the first report describing a Tunisian HLD5 patient with compound heterozygous mutations (c.521C > A (p.Ala174Glu) and c.652C > G (p.Gln218Glu)) in HYCC1 gene.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Karalok ZS, Gurkasb E, Aydinc K, Ceylaner S. Hypomyelination and congenital cataract: three siblings presentation. J Pediatr Neurosci. 2020;15:270–3.
Zara F, Biancheri R, Bruno C, Bordo L, Assereto S, Gazzerro E, et al. Deficiency of hyccin, a newly identified membrane protein, causes hypomyelination and congenital cataract. Nat Genet. 2006;38:1111–3.
Biancheri R, Zara F, Bruno C, Rossi A, Bordo L, Gazzerro E, et al. Phenotypic characterization of hypomyelination and congenital cataract. Ann Neurol. 2007;62:121–7.
Gazzerro E, Baldassari S, Giacomini C, Musante V, Fruscione F, Padula VL, et al. Hyccin, the molecule mutated in the leukodystrophy hypomyelination and congenital cataract (HCC), is a neuronal protein. PLoS ONE. 2012;7:e32180.
Baskin JM, Wu X, Christiano R, Oh MS, Schauder CM, Gazzerro E, et al. The leukodystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane. Nat Cell Biol. 2016;18:132–8.
Wolf NI, Biancheri R, Zara F, Bruno C, Gazzerro E, Rossi A, et al. Hypomyelination and congenital cataract. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJ, et al., editors. GeneReviews®. Seattle (WA): University of Washington, Seattle; 1993. http://www.ncbi.nlm.nih.gov/books/NBK2587/.
Rossi A, Biancheri R, Zara F, Bruno C, Uziel G, van der Knaap MS, et al. Hypomyelination and congenital cataract: neuroimaging features of a novel inherited white matter disorder. Am J Neuroradiol. 2008;29:301–5.
Steenweg ME, Vanderver A, Blaser S, Bizzi A, de Koning TJ, Mancini GMS, et al. Magnetic resonance imaging pattern recognition in hypomyelinating disorders. Brain. 2010;133:2971–82.
Lewin HA, Stewart-Haynes JA. A simple method for DNA extraction from leukocytes for use in PCR. BioTechniques. 1992;13:522–4.
Lorenz R, Bernhart S, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler P, et al. ViennaRNA package 2.0. Algorithms Mol Biol. 2011;6:26.
Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67.
Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N, Ashkenazy H, et al. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–33.
Zhang D, Dai L, Zhou Z, Hu J, Bai Y, Guo H. Homozygosity mapping and whole exome sequencing reveal a novel ERCC8 mutation in a Chinese consanguineous family with unique cerebellar ataxia. Clin Chim Acta. 2019;494:64–70.
Alirezaie N, Kernohan KD, Hartley T, Majewski J, Hocking TD. ClinPred: prediction tool to identify disease-relevant nonsynonymous single-nucleotide variants. Am J Hum Genet. 2018;103:474–83.
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;76:7.20.1–41.
Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–74.
Buß O, Rudat J, Ochsenreither K. FoldX as protein engineering tool: better than random based approaches? Comput Struct Biotechnol J. 2018;16:25–33.
Paladin L, Piovesan D, Tosatto SCE. SODA: prediction of protein solubility from disorder and aggregation propensity. Nucleic Acids Res. 2017;45:W236–40.
Källberg M, Margaryan G, Wang S, Ma J, Xu J. RaptorX server: a resource for template-based protein structure modeling. In: Kihara D, editor. Protein structure prediction (Methods in molecular biology). New York, NY: Springer; 2014. pp. 17–27. https://doi.org/10.1007/978-1-4939-0366-5_2.
Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23.
Traverso M, Yuregir OO, Mimouni-Bloch A, Rossi A, Aslan H, Gazzerro E, et al. Hypomyelination and congenital cataract: Identification of novel mutations in two unrelated families. Eur J Paediatr Neurol. 2013;17:108–11.
Dhaunchak AS, Colman DR, Nave KA. Misalignment of PLP/DM20 transmembrane domains determines protein misfolding in Pelizaeus–Merzbacher disease. J Neurosci. 2011;31:14961–71.
Liu N, Yamauchi J, Shooter EM. Recessive, but not dominant, mutations in peripheral myelin protein 22 gene show unique patterns of aggregation and intracellular trafficking. Neurobiol Dis. 2004;17:300–9.
Miyamoto Y, Torii T, Eguchi T, Nakamura K, Tanoue A, Yamauchi J. Hypomyelinating leukodystrophy-associated missense mutant of FAM126A/hyccin/DRCTNNB1A aggregates in the endoplasmic reticulum. J Clin Neurosci. 2014;21:1033–9.
Southwood CM, Garbern J, Jiang W, Gow A. The unfolded protein response modulates disease severity in Pelizaeus-Merzbacher disease. Neuron. 2002;36:585–96.
Dionnet E, Defour A, Da Silva N, Salvi A, Lévy N, Krahn M, et al. Splicing impact of deep exonic missense variants in CAPN3 explored systematically by minigene functional assay. Hum Mutat. 2020;41:1797–810.
Sundaresan Y, Yacoub S, Kodati B, Amankwa CE, Raola A, Zode G. Therapeutic applications of CRISPR/Cas9 gene editing technology for the treatment of ocular diseases. FEBS J. 2023;290:5248–69.
Vaz-Drago R, Custódio N, Carmo-Fonseca M. Deep intronic mutations and human disease. Hum Genet. 2017;136:1093–111.
Kraoua I, Bouyacoub Y, Drissi C, Chargui M, Rebai I, Chebil A, et al. Hypomyelination and congenital cataract: clinical, imaging, and genetic findings in three Tunisian families and literature review. Neuropediatrics. 2021;52:302–9.
Biancheri R, Zara F, Rossi A, Mathot M, Nassogne MC, Yalcinkaya C, et al. Hypomyelination and congenital cataract: broadening the clinical phenotype. Arch Neurol. 2011;68:1191.
Troncoso M, Balut F, Witting S, Rubilar C, Carrera J, Cartes F, et al. Hypomyelination and Congenital Cataract: Identification of a Novel likely pathogenic c.414+1G>A in FAM126A gene Variant. Clin. Case Rep. 2021;9:e04171.
Ugur SA, Tolun A. A deletion in DRCTNNB1A associated with hypomyelination and juvenile onset cataract. Eur J Hum Genet. 2008;16:261–4.
Acknowledgements
We thank patients and her family for their cooperation in the present study. We also thank the doctors of NeuroPediatrics Service in the CHU Hedi Chaker of Sfax for their help and availability.
Funding
This work was supported by the Ministry of Higher Education and the Scientific Research in Tunisia.
Author information
Authors and Affiliations
Contributions
Abir Ben Issa: conceptualization, methodology, software, formal analysis, and writing the original draft. Fatma Kamoun: clinical investigations. Boudour Khabou: software and bioinformatic analysis. Wafa Bouchaala: clinical review and editing. Faiza Fakhfakh: revised the final version of the manuscript. Chahnez Triki: clinical investigations.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was conducted in accordance with the principles stated in the Declaration of Helsinki-Ethical Principles for Medical Research Involving Human Subjects, Helsinki, Finland, 1964, and as amended in Fortaleza, Brazil, 2013. The study design was approved by the committee on research ethics of the University of Sfax, Tunisia.
Consent to participate
Informed consent for publication of this study was obtained from all patients and/or families involved.
Consent for publication
We certify that all contributors have read and approved the submission to this journal and that there is no financial or commercial involvement, or other conflict of interest by any author.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ben Issa, A., Kamoun, F., Khabou, B. et al. First description of novel compound heterozygous mutations in HYCC1: clinical evaluations and molecular analysis in patient with hypomyelinating leukodystrophy-5 with retrospective view. J Hum Genet 70, 75–85 (2025). https://doi.org/10.1038/s10038-024-01300-2
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-024-01300-2