Abstract
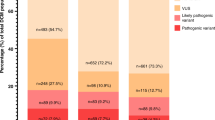
Besides the ClinGen’s efforts to standardize the ACMG/AMP criteria and European initiatives aimed at monitoring quality standards, molecular diagnostics of hereditary cardiomyopathies and heart rhythm disorders (HCHRDs) remains strongly influenced by the local strategies developed to overcome the variables in which genetic testing is requested. This is a monocentric study on the clinical and molecular findings of 363 pedigrees with various HCHRDs. ACMG/AMP criteria were adapted according to the ClinGen’s material and internal specifications. Phenotypes were reviewed according to known disease-gene associations and the concurrence of multiple variants in the same individual. Relatives were studied when available and the significance of selected variants was supported by RNA- studies before reporting. One or more (likely) pathogenic variants were found in 80 pedigrees (22.0%), while 96 (26.4%) displayed one or more variants of uncertain significance (VUS) only. The 132 identified VUS were sub-tiered according to the Bayesian score in three categories presenting distinguishable patterns of selected criteria. VUS_high showed profiles of key molecular criteria and resembled deleterious variants according to the combinations of assigned criteria, while the VUS_low category displayed a high chance of conflicting combinations of criteria and unsupported disease-gene associations. Reclassification to likely pathogenic by the application of applicable clinical criteria (PVS1_Strength, PP1 and PP4) was accessible to VUS_high and a few VUS_mid only. This work supports the combined need to (i) introduce VUS sub-tiering, (ii) consider known disease-gene associations, (iii) stringently apply clinical criteria and (iv) incorporate RNA data to improve the clinical significance of genetic testing in HCHRDs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Ommen SR, Mital S, Burke MA, Day SM, Deswal A, Elliott P, et al. 2020 AHA/ACC Guideline for the diagnosis and treatment of patients with hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation. 2020;142:e533–e57. https://doi.org/10.1161/CIR.0000000000000938.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. https://doi.org/10.1038/gim.2015.30.
Arbustini E, Behr ER, Carrier L, van Duijn C, Evans P, Favalli V, et al. Interpretation and actionability of genetic variants in cardiomyopathies: a position statement from the European Society of Cardiology Council on cardiovascular genomics. Eur Heart J. 2022;43:1901–16. https://doi.org/10.1093/eurheartj/ehab895.
Kelly MA, Caleshu C, Morales A, Buchan J, Wolf Z, Harrison SM, et al. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet Med. 2018;20:351–9. https://doi.org/10.1038/gim.2017.218.
Ingles J, Goldstein J, Thaxton C, Caleshu C, Corty EW, Crowley SB, et al. Evaluating the clinical validity of hypertrophic cardiomyopathy genes. Circ Genom Precis Med. 2019;12:e002460. https://doi.org/10.1161/CIRCGEN.119.002460.
James CA, Jongbloed JDH, Hershberger RE, Morales A, Judge DP, Syrris P, et al. International evidence based reappraisal of genes associated with arrhythmogenic right ventricular cardiomyopathy using the clinical genome resource framework. Circ Genom Precis Med. 2021;14:e003273. https://doi.org/10.1161/CIRCGEN.120.003273.
Jordan E, Peterson L, Ai T, Asatryan B, Bronicki L, Brown E, et al. Evidence-based assessment of genes in dilated cardiomyopathy. Circulation. 2021;144:7–19. https://doi.org/10.1161/CIRCULATIONAHA.120.053033.
Hayesmoore JB, Bhuiyan ZA, Coviello DA, du Sart D, Edwards M, Iascone M, et al. EMQN: Recommendations for genetic testing in inherited cardiomyopathies and arrhythmias. Eur J Hum Genet. 2023;31:1003–9. https://doi.org/10.1038/s41431-023-01421-w.
Lopes LR, Ho CY, Elliott PM. Genetics of hypertrophic cardiomyopathy: established and emerging implications for clinical practice. Eur Heart J. 2024;45:2727–34. https://doi.org/10.1093/eurheartj/ehae421.
Porretta AP, Probst V, Bhuiyan ZA, Davoine E, Deliniere A, Pascale P, et al. SCN5A overlap syndromes: An open-minded approach. Heart Rhythm. 2022;19:1363–8. https://doi.org/10.1016/j.hrthm.2022.03.1223.
Kolokotronis K, Kuhnisch J, Klopocki E, Dartsch J, Rost S, Huculak C, et al. Biallelic mutation in MYH7 and MYBPC3 leads to severe cardiomyopathy with left ventricular noncompaction phenotype. Hum Mutat. 2019;40:1101–14. https://doi.org/10.1002/humu.23757.
Rosario KF, Karra R, Amos K, Landstrom AP, Lakdawala NK, Brezitski K, et al. LMNA cardiomyopathy: important considerations for the heart failure clinician. J Card Fail. 2023;29:1657–66. https://doi.org/10.1016/j.cardfail.2023.08.016.
Xu T, Yang Z, Vatta M, Rampazzo A, Beffagna G, Pilichou K, et al. Compound and digenic heterozygosity contributes to arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol. 2010;55:587–97. https://doi.org/10.1016/j.jacc.2009.11.020.
Tavtigian SV, Greenblatt MS, Harrison SM, Nussbaum RL, Prabhu SA, Boucher KM, et al. Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet Med. 2018;20:1054–60. https://doi.org/10.1038/gim.2017.210.
Josephs KS, Roberts AM, Theotokis P, Walsh R, Ostrowski PJ, Edwards M, et al. Beyond gene-disease validity: capturing structured data on inheritance, allelic requirement, disease-relevant variant classes, and disease mechanism for inherited cardiac conditions. Genome Med. 2023;15:86. https://doi.org/10.1186/s13073-023-01246-8.
Olivotto I, Girolami F, Ackerman MJ, Nistri S, Bos JM, Zachara E, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2008;83:630–8. https://doi.org/10.4065/83.6.630.
Page SP, Kounas S, Syrris P, Christiansen M, Frank-Hansen R, Andersen PS, et al. Cardiac myosin binding protein-C mutations in families with hypertrophic cardiomyopathy: disease expression in relation to age, gender, and long term outcome. Circ Cardiovasc Genet. 2012;5:156–66. https://doi.org/10.1161/CIRCGENETICS.111.960831.
Roberts AM, DiStefano MT, Riggs ER, Josephs KS, Alkuraya FS, Amberger J, et al. Toward robust clinical genome interpretation: Developing a consistent terminology to characterize Mendelian disease-gene relationships-allelic requirement, inheritance modes, and disease mechanisms. Genet Med. 2024;26:101029. https://doi.org/10.1016/j.gim.2023.101029.
Miller DT, Lee K, Abul-Husn NS, Amendola LM, Brothers K, Chung WK, et al. ACMG SF v3.2 list for reporting of secondary findings in clinical exome and genome sequencing: A policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2023;25:100866. https://doi.org/10.1016/j.gim.2023.100866.
Houge G, Laner A, Cirak S, de Leeuw N, Scheffer H, den Dunnen JT. Stepwise ABC system for classification of any type of genetic variant. Eur J Hum Genet. 2022;30:150–9. https://doi.org/10.1038/s41431-021-00903-z.
Rehm HL, Alaimo JT, Aradhya S, Bayrak-Toydemir P, Best H, Brandon R, et al. The landscape of reported VUS in multi-gene panel and genomic testing: Time for a change. Genet Med. 2023;25:100947. https://doi.org/10.1016/j.gim.2023.100947.
Chen E, Facio FM, Aradhya KW, Rojahn S, Hatchell KE, Aguilar S, et al. Rates and classification of variants of uncertain significance in hereditary disease genetic testing. JAMA Netw Open. 2023;6:e2339571 https://doi.org/10.1001/jamanetworkopen.2023.39571.
Acknowledgements
The authors would like to thank all the participants who donated valuable samples and data to allow this study.
Funding
This work was funded by the Italian Ministry of Health (Ricerca Corrente 2022-2024 and PNRR-MCNT2-2023-12377305 “EUCARDIS”) to MC, and by Fondazione Telethon Core Grant, Armenise-Harvard Foundation Career Development Award, European Research Council (grant agreement 759154, CellKarma), Italian Ministry of Health (Piano Operativo Salute Traiettoria 3, “Genomed”; Ricerca Finalizzata 2021, “genOMICA”; PNRR-MCNT2-2023-12377305 “EUCARDIS”) and Italian Ministry of University and Research (Progetti di Rilevante Interesse Nazionale 2022) to DC.
Author information
Authors and Affiliations
Contributions
Conceptualization M.C., S.M., G.D., S.M., C.F.; methodology G.N., A.F., M.C., C.F., E.D., P.P., M.P.L,; validation G.N.,C.F., R.P., O.P., M.C., D.C.; clinical evaluation A.P., D.R.P., M.C., G.D.L., C.C., R.S.M., R.D.S., L.V.; writing—original draft preparation M.C.,S.M., G.D., F.C.; supervision M.C. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
Davide Cacchiarelli is founder, shareholder, and consultant of NEGEDIA S.r.l. All the other authors declare that there is no conflict of interest concerning this work.
Ethics approval
This study is in accordance with the 1984 Helsinki declaration and subsequent revisions, and received IRB approval by the local ethics committee ‘Azienda Ospedaliero Universitaria Policlinico Foggia’ (approval no. 89/CE/2024).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Castori, M., Mastroianno, S., Fontana, A. et al. Variant sub-tiering, disease-gene associations and strictness of clinical criteria improves the interpretation of variants of uncertain significance in hereditary cardiomyopathies and rhythm disorders. J Hum Genet 70, 349–358 (2025). https://doi.org/10.1038/s10038-025-01344-y
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s10038-025-01344-y