Abstract
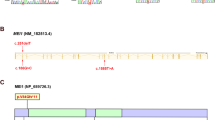
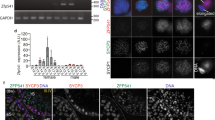
Non-obstructive azoospermia (NOA) represents the severe form of male infertility, affecting approximately 1% of men during their reproductive years. It is marked by the absence of sperm production caused by testicular dysfunction and has many genetic origins. However, the genetic factors underlying most NOA cases are still unclear. Meiosis, a crucial process ensuring accurate chromosome segregation and generating genetic diversity in gametes, is susceptible to genetic disruptions that may result in NOA. In this study, whole exome sequencing (WES) was conducted on 969 NOA patients, identifying six compound heterozygous KCTD19 variants in three Chinese pedigrees. KCTD19 has been demonstrated to interact with ZFP541 and HDAC1, thereby participating in the modulation of chromatin remodeling and transcriptional programs during meiosis in mice. Herein, our findings expand the phenotypic and mutational spectrum of KCTD19 in male infertility and provide further insights into its role during meiosis. This research underscores the importance of KCTD19 in meiotic progression and male fertility, highlighting the need for further investigation into the molecular mechanisms underlying gametogenic failure in NOA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Wosnitzer M, Goldstein M, Hardy MP. Review of azoospermia. Spermatogenesis. 2014;4:e28218.
Tournaye H, Krausz C, Oates RD. Novel concepts in the aetiology of male reproductive impairment. Lancet Diabetes Endocrinol. 2017;5:544–53.
Corona G, Minhas S, Giwercman A, Bettocchi C, Dinkelman-Smit M, Dohle G, et al. Sperm recovery and ICSI outcomes in men with non-obstructive azoospermia: a systematic review and meta-analysis. Hum Reprod Update. 2019;25:733–57.
Hu Z, Li Z, Yu J, Tong C, Lin Y, Guo X, et al. Association analysis identifies new risk loci for non-obstructive azoospermia in Chinese men. Nat Commun. 2014;5:3857.
Grey C, de Massy B. Chromosome organization in early meiotic prophase. Front Cell Dev Biol. 2021;9:688878.
Borde V, de Massy B. Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr Opin Genet Dev. 2013;23:147–55.
Xie C, Wang W, Tu C, Meng L, Lu G, Lin G, et al. Meiotic recombination: insights into its mechanisms and its role in human reproduction with a special focus on non-obstructive azoospermia. Hum Reprod Update. 2022;28:763–97.
Bolcun-Filas E, Handel MA. Meiosis: the chromosomal foundation of reproduction. Biol Reprod. 2018;99:112–26.
He WB, Tu CF, Liu Q, Meng LL, Yuan SM, Luo AX, et al. DMC1 mutation that causes human non-obstructive azoospermia and premature ovarian insufficiency identified by whole-exome sequencing. J Med Genet. 2018;55:198–204.
Li P, Ji Z, Zhi E, Zhang Y, Han S, Zhao L, et al. Novel bi-allelic MSH4 variants causes meiotic arrest and non-obstructive azoospermia. Reprod Biol Endocrinol. 2022;20:21.
Ji Z, Yao C, Yang C, Huang C, Zhao L, Han X, et al. Novel hemizygous mutations of TEX11 cause meiotic arrest and non-obstructive azoospermia in Chinese Han population. Front Genet. 2021;12:741355.
Xu S, Zhao J, Gao F, Zhang Y, Luo J, Zhang C, et al. A bi-allelic REC114 loss-of-function variant causes meiotic arrest and nonobstructive azoospermia. Clin Genet. 2024;105:440–5.
Zhao J, Ji Z, Meng G, Luo J, Zhang Y, Ou N, et al. Identification of a missense variant of MND1 in meiotic arrest and non-obstructive azoospermia. J Hum Genet. 2023;68:729–35.
Tang D, Lv M, Gao Y, Cheng H, Li K, Xu C, et al. Novel variants in helicase for meiosis 1 lead to male infertility due to non-obstructive azoospermia. Reprod Biol Endocrinol. 2021;19:129.
Yao C, Yang C, Zhao L, Li P, Tian R, Chen H, et al. Bi-allelic SHOC1 loss-of-function mutations cause meiotic arrest and non-obstructive azoospermia. J Med Genet. 2021;58:679–86.
Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol. 2018;15:369–84.
Choi E, Han C, Park I, Lee B, Jin S, Choi H, et al. A novel germ cell-specific protein, SHIP1, forms a complex with chromatin remodeling activity during spermatogenesis. J Biol Chem. 2008;283:35283–94.
Horisawa-Takada Y, Kodera C, Takemoto K, Sakashita A, Horisawa K, Maeda R, et al. Meiosis-specific ZFP541 repressor complex promotes developmental progression of meiotic prophase towards completion during mouse spermatogenesis. Nat Commun. 2021;12:3184.
Oura S, Koyano T, Kodera C, Horisawa-Takada Y, Matsuyama M, Ishiguro KI, et al. KCTD19 and its associated protein ZFP541 are independently essential for meiosis in male mice. PLoS Genet. 2021;17:e1009412.
Fang K, Li Q, Wei Y, Zhou C, Guo W, Shen J, et al. Prediction and validation of mouse meiosis-essential genes based on spermatogenesis proteome dynamics. Mol Cell Proteom. 2021;20:100014.
Zhang Y, Huang X, Xu Q, Yu M, Shu M, Shan S, et al. Homozygous nonsense variants of KCTD19 cause male infertility in humans and mice. J Genet Genomics. 2023;50:615–9.
Liu J, Rahim F, Zhou J, Fan S, Jiang H, Yu C, et al. Loss-of-function variants in KCTD19 cause non-obstructive azoospermia in humans. iScience. 2023;26:107193.
Wang W, Su L, Meng L, He J, Tan C, Yi D, et al. Biallelic variants in KCTD19 associated with male factor infertility and oligoasthenoteratozoospermia. Hum Reprod. 2023;38:1399–411.
Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HW, Behre HM, et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update. 2010;16:231–45.
Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82.
Kishore S, Khanna A, Stamm S. Rapid generation of splicing reporters with pSpliceExpress. Gene. 2008;427:104–10.
Li Y, Meng R, Li S, Gu B, Xu X, Zhang H, et al. The ZFP541-KCTD19 complex is essential for pachytene progression by activating meiotic genes during mouse spermatogenesis. J Genet Genomics. 2022;49:1029–41.
Pinkas DM, Sanvitale CE, Bufton JC, Sorrell FJ, Solcan N, Chalk R, et al. Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases. Biochem J. 2017;474:3747–61.
Ahmad KF, Melnick A, Lax S, Bouchard D, Liu J, Kiang CL, et al. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12:1551–64.
Minor DL, Lin YF, Mobley BC, Avelar A, Jan YN, Jan LY, et al. The polar T1 interface is linked to conformational changes that open the voltage-gated potassium channel. Cell. 2000;102:657–70.
Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 2004;23:1681–7.
Faqeih EA, Almannai M, Saleh MM, AlWadei AH, Samman MM, Alkuraya FS. Phenotypic characterization of KCTD3-related developmental epileptic encephalopathy. Clin Genet. 2018;93:1081–6.
Metz KA, Teng X, Coppens I, Lamb HM, Wagner BE, Rosenfeld JA, et al. KCTD7 deficiency defines a distinct neurodegenerative disorder with a conserved autophagy-lysosome defect. Ann Neurol. 2018;84:766–80.
Teng X, Aouacheria A, Lionnard L, Metz KA, Soane L, Kamiya A, et al. KCTD: a new gene family involved in neurodevelopmental and neuropsychiatric disorders. CNS Neurosci Ther. 2019;25:887–902.
Zhou X, Fang K, Liu Y, Li W, Tan Y, Zhang J, et al. ZFP541 and KCTD19 regulate chromatin organization and transcription programs for male meiotic progression. Cell Prolif. 2024;57:e13567.
Xu J, Gao J, Liu J, Huang X, Zhang H, Ma A, et al. ZFP541 maintains the repression of pre-pachytene transcriptional programs and promotes male meiosis progression. Cell Rep. 2022;38:110540.
Acknowledgements
The authors express their gratitude to the participating patient’s family for their invaluable contribution to this study.
Funding
This study was supported by National Key Research and Development Program of China (2022YFC2702701), National Natural Science Foundation of China (82401869, 82371616), Inner Mongolia Academy of Medical Sciences Public Hospital Joint Science and Technology Project (2023GLLH0045), Specific Project of Shanghai Jiao Tong University for “Invigorating Inner Mongolia through Science and Technology”(2022XYJG001-01-19), and Scientific Research Startup Funding for High-Level Talents of Taizhou School of Clinical Medicine (TZKY2023RC01).
Author information
Authors and Affiliations
Contributions
All the authors participated in the development and design of the study. YXZ, NL, and ZL designed the research; SX and CWZ performed the research; Clinical data were collected by CWZ, WZN, DWQ, and ZZM. PL, YHH, JXZ, CZD and FRB collected and prepared clinical samples. SX, CCY and YFS performed the bioinformatics analysis; JXZ, ZZ, NL and YXZ analyzed the data; SX, CWZ, CCY and YXZ wrote and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All participants gave written informed consent to take part in the study. The research was approved by the Institutional Ethical Review Committee of Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine (license number 2022SQ294). Consent was obtained from the donors for the use of clinical data and testicular tissue in research.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, S., Zhang, C., Yao, C. et al. Bi-allelic KCTD19 variants associated with meiotic arrest and non-obstructive azoospermia in humans. J Hum Genet 70, 427–437 (2025). https://doi.org/10.1038/s10038-025-01350-0
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s10038-025-01350-0