Abstract
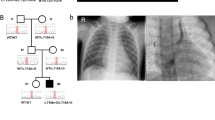
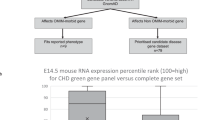
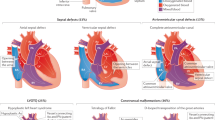
Congenital heart disease (CHD) affects approximately 1% of liveborn infants. Among primary ciliary dyskinesia (PCD) cases, about 50% present with situs inversus totalis, and 6.3% have heterotaxy with CHD. The incidence of CHD is significantly higher in heterotaxy patients compared to the general population (57% vs. 1%). However, comprehensive studies on CHD related to laterality defects are still limited. In this study, we retrospectively analyzed 18,781 CHD patients to determine the prevalence of laterality defects. To evaluate the association between specific complex CHD phenotypes and laterality defects, we utilized a binary logistic regression model. Additionally, we performed whole-exome sequencing (WES) on 121 CHD patients with laterality defects. The results showed that 1.1% of CHD patients had laterality defects (206/18,781), with 0.4% presenting as situs inversus totalis and 0.7% as situs ambiguus. The prevalence of laterality defects was higher in complex CHD cases (5.4%) compared to simple CHD (0.4%). Notably, single atrium with single ventricle (SA+SV) was strongly associated with laterality defects (OR = 48.23, p < 0.001). Among the 121 CHD patients with situs abnormalities, WES identified pathogenic gene variants in 13.2%, with 9.1% harboring known pathogenic genes (ZIC3, NODAL, NKX2-5, GDF1, MMP21, PKD1L1, CCDC151, DNAAF4, LRRC56) and 4.1% exhibiting variants in candidate genes (FMNL3, C1ORF127, CFAP157, C10ORF107, MYO1D). This study revealed both established and novel gene candidates, contributing to our understanding of the genetic basis of laterality defects in CHD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data are available upon reasonable request from the corresponding author.
References
Van Der Bom T, Zomer AC, Zwinderman AH, Meijboom FJ, Bouma BJ, Mulder BJ. The changing epidemiology of congenital heart disease. Nat Rev Cardiol. 2011;8:50–60.
Van Der Bom T, Bouma BJ, Meijboom FJ, Zwinderman AH, Mulder BJ. The prevalence of adult congenital heart disease, results from a systematic review and evidence based calculation. Am Heart J. 2012;164:568–75.
Newburger JW, Sleeper LA, Bellinger DC, Goldberg CS, Tabbutt S, Lu M, et al. Early developmental outcome in children with hypoplastic left heart syndrome and related anomalies: the single ventricle reconstruction trial. Circulation. 2012;125:2081–91.
Swisher M, Jonas R, Tian X, Lee ES, Lo CW, Leatherbury L. Increased postoperative and respiratory complications in patients with congenital heart disease associated with heterotaxy. J Thorac Cardiovasc Surg. 2011;141:637–44.
Antony D, et al. Spectrum of Genetic Variants in a Cohort of 37 Laterality Defect Cases. Front Genet. 2022;13:861236.
Noone PG, Leigh MW, Sannuti A, Minnix SL, Carson JL, Hazucha M, et al. Primary ciliary dyskinesia: diagnostic and phenotypic features. Am J Respir Crit Care Med. 2004;169:459–67.
Kennedy MP, Omran H, Leigh MW, Dell S, Morgan L, Molina PL, et al. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation. 2007;115:2814–21.
Shapiro AJ, Davis SD, Ferkol T, Dell SD, Rosenfeld M, Olivier KN, et al. Laterality defects other than situs inversus totalis in primary ciliary dyskinesia: insights into situs ambiguus and heterotaxy. Chest. 2014;146:1176–86.
Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521:520–4.
Francis RJ, Christopher A, Devine WA, Ostrowski L, Lo C. Congenital heart disease and the specification of left-right asymmetry. Am J Physiol Heart Circ Physiol. 2012;302:H2102–H11.
Marelli AJ, Mackie AS, Ionescu-Ittu R, Rahme E, Pilote L. Congenital heart disease in the general population: changing prevalence and age distribution. Circulation. 2007;115:163–72.
Guo T, Tan Z-P, Chen H-M, Zheng DY, Liu L, Huang XG, et al. An effective combination of whole-exome sequencing and runs of homozygosity for the diagnosis of primary ciliary dyskinesia in consanguineous families. Scientific Rep. 2017;7:7905.
Zhang W, Li D, Wei S, Guo T, Wang J, Luo H, et al. Correction: Whole-exome sequencing identifies a novel CCDC151 mutation, c.325GT (p.E109X), in a patient with primary ciliary dyskinesia and situs inversus. J Hum Genet. 2019;64:829.
Alsamri MT, Alabdouli A, Iram D, Alkalbani AM, Almarzooqi AS, Souid AK, et al. A Study on the Genetics of Primary Ciliary Dyskinesia. J Clin Med. 2021;10:5102.
Nakhleh N, Francis R, Giese RA, Tian X, Li Y, Zariwala MA, et al. High prevalence of respiratory ciliary dysfunction in congenital heart disease patients with heterotaxy. Circulation. 2012;125:2232–42.
Lin AE, Ticho BS, Houde K, Westgate MN, Holmes LB. Heterotaxy: associated conditions and hospital-based prevalence in newborns. Genet Med. 2000;2:157–72.
Zhu L, Belmont JW, Ware SM. Genetics of human heterotaxias. European J Hum Genet: EJHG. 2006;14:17–25.
Hashmi A, Abu-Sulaiman R, Mccrindle BW, Smallhorn JF, Williams WG, Freedom RM. Management and outcomes of right atrial isomerism: a 26-year experience. J Am Coll Cardiol. 1998;31:1120–6.
Taketazu M, Lougheed J, Yoo S-J, Lim JS, Hornberger LK. Spectrum of cardiovascular disease, accuracy of diagnosis, and outcome in fetal heterotaxy syndrome. Am J Cardiol. 2006;97:720–4.
Gilljam T, Mccrindle BW, Smallhorn JF, Williams WG, Freedom RM. Outcomes of left atrial isomerism over a 28-year period at a single institution. J Am Coll Cardiol. 2000;36:908–16.
Bartram U, Wirbelauer J, Speer CP. Heterotaxy syndrome - asplenia and polysplenia as indicators of visceral malposition and complex congenital heart disease. Biol Neonate. 2005;88:278–90.
Hoffman JIE, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–900.
Shiratori H, Hamada H. The left-right axis in the mouse: from origin to morphology. Development (Camb, Engl). 2006;133:2095–104.
Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–8.
Bisgrove BW, Morelli SH, Yost HJ. Genetics of human laterality disorders: insights from vertebrate model systems. Annu Rev Genomics Hum Genet. 2003;4:1–32.
Ware SM, Peng J, Zhu L, Fernbach S, Colicos S, Casey B, et al. Identification and functional analysis of ZIC3 mutations in heterotaxy and related congenital heart defects. American J Hum Genet. 2004;74:93–105.
Sifrim A, Hitz M-P, Wilsdon A, Breckpot J, Turki SH, Thienpont B, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nature Genet. 2016;48:1060–5.
Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nature Genet. 2017;49:1593–601.
Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–6.
Snarr BS, O'neal JL, Chintalapudi MR, Wirrig EE, Phelps AL, Kubalak SW, et al. Isl1 expression at the venous pole identifies a novel role for the second heart field in cardiac development. Circulation Res. 2007;101:971–4.
Campione M, Ros MA, Icardo JM, Piedra E, Christoffels VM, Schweickert A, et al. Pitx2 expression defines a left cardiac lineage of cells: evidence for atrial and ventricular molecular isomerism in the iv/iv mice. Dev Biol. 2001;231:252–64.
Romanowicz J, Sinha P, Donofrio MT, Schidlow DN. Predicting cardiac anatomy, physiology, and surgical management based on fetal echocardiography in heterotaxy syndrome. Am J Perinatol. 2021;40:1081–7.
Harrison MJ, Shapiro AJ, Kennedy MP. Congenital heart disease and primary ciliary dyskinesia. Paediatr Respir Rev. 2016;18:25–32.
Williams K, Carson J, Lo C. Genetics of congenital heart disease. Biomolecules. 2019;9:879.
Djenoune L, Berg K, Brueckner M, Yuan S. A change of heart: new roles for cilia in cardiac development and disease. Nat Rev Cardiol. 2022;19:211–27.
Gabriel GC, Young CB, Lo CW. Role of cilia in the pathogenesis of congenital heart disease. Semin Cell Dev Biol. 2021;110:2–10.
Chung B, Shaffer LG, keating S, Johnson J, Casey B, Chitayat D. From VACTERL-H to heterotaxy: variable expressivity of ZIC3-related disorders. American J Med Genet Part A. 2011;155A:1123–8.
Purandare SM, Ware SM, Kwan KM, Gebbia M, Bassi MT, Deng JM, et al. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development (Camb, Engl). 2002;129:2293–302.
Chung I-M, Rajakumar G. Genetics of Congenital Heart Defects: The NKX2-5 Gene, a Key Player. Genes (Basel). 2016;7:6.
Gao X, Zheng P, Yang L, Luo H, Zhang C, Qiu Y, et al. Association of functional variant in GDF1 promoter with risk of congenital heart disease and its regulation by Nkx2.5. Clin Sci (Lond). 2019;133:1281–95.
Yuan ZZ, Fan LL, Jiang ZC, Yang YF, Tan ZP. A novel nonsense MMP21 variant causes dextrocardia and congenital heart disease in a han chinese patient. Front Cardiovasc Med. 2020;7:582350.
Bonnefoy S, Watson CM, Kernohan KD, Lemos M, Hutchinson S, Poulter JA, et al. Biallelic mutations in LRRC56, encoding a protein associated with intraflagellar transport, cause mucociliary clearance and laterality defects. American J Hum Genet. 2018;103:727–39.
Hu S, Dasbiswas K, Guo Z, Tee YH, Thiagarajan V, Hersen P, et al. Long-range self-organization of cytoskeletal myosin II filament stacks. Nat Cell Biol. 2017;19:133–41.
Phng L-K, Gebala V, Bentley K, Philippides A, Wacker A, Mathivet T, et al. Formin-mediated actin polymerization at endothelial junctions is required for vessel lumen formation and stabilization. Dev Cell. 2015;32:123–32.
Weidemann M, Schuster-Gossler K, Stauber M, Wrede C, Hegermann J, Ott T, et al. CFAP157 is a murine downstream effector of FOXJ1 that is specifically required for flagellum morphogenesis and sperm motility. Development (Camb, Engl). 2016;143:4736–48.
Kawashima A, Kigoshi T, Katoh Y, Ishikawa Y, Shawki HH, Inoue N, et al. CABCOCO1, a novel coiled-coil protein With calcium-binding activity, is localized in the sperm flagellum. Mol Reprod Dev. 2016;83:912–26.
Juan T, Géminard C, Coutelis J-B, Cerezo D, Polès S, Noselli S, et al. Myosin1D is an evolutionarily conserved regulator of animal left-right asymmetry. Nature Commun. 2018;9:1942.
Yuan Z, Zhu X, Xie X, Wang C, Gu H, Yang J, et al. Identification of a novel MYO1D variant associated with laterality defects, congenital heart diseases, and sperm defects in humans. Front Med. 2024;18:558–64.
Acknowledgements
The authors express their sincere gratitude to all patients and their families for their invaluable support and cooperation.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81970268, to Dr. Tan) and Natural Science Foundation of Hunan Provincial (No.2023JJ30781, to Dr. Tan).
Author information
Authors and Affiliations
Contributions
Z-PT designed the overall study and performed data analysis. X-HX processed the WES data, validated the mutation, and drafted the manuscript. Y-FY, J-LC, L-X and W-ZZ enrolled the subjects. H-G, J-LY, K-LQ carried out the clinical data analysis. Z-ZY analyzed the data and edited the manuscript. All authors read and approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics statement
The study involving human subjects were reviewed and approved by Ethics Committee of the Second Xiangya Hospital of the Central South University (Approval No. 322, 2021). A signed written informed consent was obtained from the legal guardians of the patient for the publication of any potentially identifiable images or data included in this article.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xie, Xh., Gu, H., Yuan, Zz. et al. The prevalence of laterality defects in patients with congenital heart disease. J Hum Genet 70, 453–461 (2025). https://doi.org/10.1038/s10038-025-01351-z
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s10038-025-01351-z