Abstract
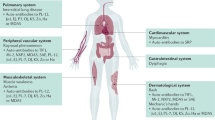
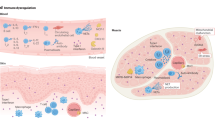
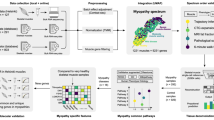
Myositis is a heterogeneous group of inflammatory muscular disorders. Although the main etiology is autoimmune chronic inflammation, the underlying pathomechanism remains unclear. Advances in genetic technology have provided important insights into its complex pathophysiology. Large genetic studies on myositis have advocated a relationship with several HLA loci and possible disease susceptibility genes in non-HLA genes. Idiopathic inflammatory myopathy, or autoimmune myositis, was originally divided into polymyositis and dermatomyositis. However, this classification has recently been revised based on updated information on the pathophysiology of autoimmune myositis. Autoimmune myositis is currently understood to include at least four major clinicopathologically distinct entities: dermatomyositis, antisynthetase syndrome, inclusion body myositis, and immune-mediated necrotizing myopathy. This review aims to consolidate knowledge of the genetics of myositis in order to meet the current classification and highlights key findings for a more detailed understanding of the underlying pathomechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Miller FW, Chen W, O’Hanlon TP, Cooper RG, Vencovsky J, Rider LG, et al. Genome-wide association study identifies HLA 8.1 ancestral haplotype alleles as major genetic risk factors for myositis phenotypes. Genes Immun. 2015;16:470–80. https://doi.org/10.1038/gene.2015.28.
Rothwell S, Cooper RG, Lundberg IE, Miller FW, Gregersen PK, Bowes J, et al. Dense genotyping of immune-related loci in idiopathic inflammatory myopathies confirms HLA alleles as the strongest genetic risk factor and suggests different genetic background for major clinical subgroups. Ann Rheum Dis. 2016;75:1558–66. https://doi.org/10.1136/annrheumdis-2015-208119.
Bohan A, Peter JB. Polymyositis and dermatomyositis (first of two parts). N Engl J Med. 1975;292:344–7. https://doi.org/10.1056/NEJM197502132920706.
Bohan A, Peter JB. Polymyositis and dermatomyositis (second of two parts). N Engl J Med. 1975;292:403–7. https://doi.org/10.1056/NEJM197502202920807.
Uruha A, Uruha S. Update on pathology of inflammatory myopathy. Clin Exp Neuroimmunol. 2025. https://doi.org/10.1111/cen3.12824.
Tanboon J, Uruha A, Stenzel W, Nishino I. Where are we moving in the classification of idiopathic inflammatory myopathies?. Curr Opin Neurol. 2020;33:590–603. https://doi.org/10.1097/WCO.0000000000000855.
Suzuki S, Uruha A, Suzuki N, Nishino I. Integrated diagnosis project for inflammatory myopathies: an association between autoantibodies and muscle pathology. Autoimmun Rev. 2017;16:693–700. https://doi.org/10.1016/j.autrev.2017.05.003.
Inoue M, Tanboon J, Hirakawa S, Komaki H, Fukushima T, Awano H, et al. Association of dermatomyositis sine dermatitis with anti-nuclear matrix protein 2 autoantibodies. JAMA Neurol. 2020;77:872–7. https://doi.org/10.1001/jamaneurol.2020.0673.
Noguchi E, Uruha A, Suzuki S, Hamanaka K, Ohnuki Y, Tsugawa J, et al. Skeletal muscle involvement in antisynthetase syndrome. JAMA Neurol. 2017;74:992–9. https://doi.org/10.1001/jamaneurol.2017.0934.
Mariampillai K, Granger B, Amelin D, Guiguet M, Hachulla E, Maurier F, et al. Development of a new classification system for idiopathic inflammatory myopathies based on clinical manifestations and myositis-specific autoantibodies. JAMA Neurol. 2018;75:1528–37. https://doi.org/10.1001/jamaneurol.2018.2598.
Rothwell S, Chinoy H, Lamb JA, Miller FW, Rider LG, Wedderburn LR, et al. Focused HLA analysis in Caucasians with myositis identifies significant associations with autoantibody subgroups. Ann Rheum Dis. 2019;78:996–1002. https://doi.org/10.1136/annrheumdis-2019-215046.
Deakin CT, Bowes J, Rider LG, Miller FW, Pachman LM, Sanner H, et al. Association with HLA-DRβ1 position 37 distinguishes juvenile dermatomyositis from adult-onset myositis. Hum Mol Genet. 2022;31:2471–81. https://doi.org/10.1093/hmg/ddac019.
Gambino CM, Aiello A, Accardi G, Caruso C, Candore G. Autoimmune diseases and 8.1 ancestral haplotype: an update. HLA. 2018;92:137–43. https://doi.org/10.1111/tan.13305.
Lande A, Andersen I, Egeland T, Lie BA, Viken MK. HLA -A, -C, -B, -DRB1, -DQB1 and -DPB1 allele and haplotype frequencies in 4514 healthy Norwegians. Hum Immunol. 2018;79:527–9. https://doi.org/10.1016/j.humimm.2018.04.012.
Oyama M, Ohnuki Y, Inoue M, Uruha A, Yamashita S, Yutani S, et al. HLA-DRB1 allele and autoantibody profiles in Japanese patients with inclusion body myositis. PLoS One. 2020;15:e0237890. https://doi.org/10.1371/journal.pone.0237890.
Oyama M, Ohnuki Y, Uruha A, Saito Y, Nishimori Y, Suzuki S, et al. Association between HLA alleles and autoantibodies in dermatomyositis defined by sarcoplasmic expression of myxovirus resistance protein A. J Rheumatol. 2023;50:1159–64. https://doi.org/10.3899/jrheum.2022-1321.
Kang EH, Go DJ, Mimori T, Lee SJ, Kwon HM, Park JW, et al. Novel susceptibility alleles in HLA region for myositis and myositis specific autoantibodies in Korean patients. Semin Arthritis Rheum. 2019;49:283–7. https://doi.org/10.1016/j.semarthrit.2019.03.005.
Lamb JA. The genetics of autoimmune myositis. Front Immunol. 2022;13:886290. https://doi.org/10.3389/fimmu.2022.886290.
Kochi Y, Kamatani Y, Kondo Y, Suzuki A, Kawakami E, Hiwa R, et al. Splicing variant of WDFY4 augments MDA5 signalling and the risk of clinically amyopathic dermatomyositis. Ann Rheum Dis. 2018;77:602–11. https://doi.org/10.1136/annrheumdis-2017-212149.
Guo L, Zhang X, Pu W, Zhao J, Wang K, Zhang D, et al. WDFY4 polymorphisms in Chinese patients with anti-MDA5 dermatomyositis is associated with rapid progressive interstitial lung disease. Rheumatology. 2023;62:2320–4. https://doi.org/10.1093/rheumatology/kead006.
Lyu X, Lamb JA, Chinoy H. The clinical relevance of WDFY4 in autoimmune diseases in diverse ancestral populations. Rheumatology. 2024;63:3255–62. https://doi.org/10.1093/rheumatology/keae183.
Rothwell S, Amos CI, Miller FW, Rider LG, Lundberg IE, Gregersen PK, et al. Identification of novel associations and localization of signals in idiopathic inflammatory myopathies using genome-wide imputation. Arthritis Rheumatol. 2023;75:1021–7. https://doi.org/10.1002/art.42434.
Zhu C, Han Y, Byun J, Xiao X, Rothwell S, Miller FW, et al. Genetic architecture of idiopathic inflammatory myopathies from meta-analyses. Arthritis Rheumatol. 2025;77:750–64. https://doi.org/10.1002/art.43088.
Rothwell S, Cooper RG, Lundberg IE, Gregersen PK, Hanna MG, Machado PM, et al. Immune-array analysis in sporadic inclusion body myositis reveals HLA-DRB1 amino acid heterogeneity across the myositis spectrum. Arthritis Rheumatol. 2017;69:1090–9. https://doi.org/10.1002/art.40045.
Slater N, Sooda A, McLeish E, Beer K, Brusch A, Shakya R, et al. High-resolution HLA genotyping in inclusion body myositis refines 8.1 ancestral haplotype association to DRB1*03:01:01 and highlights pathogenic role of arginine-74 of DRβ1 chain. J Autoimmun. 2024;142:103150. https://doi.org/10.1016/j.jaut.2023.103150.
Mavroudis I, Knights M, Petridis F, Chatzikonstantinou S, Karantali E, Kazis D. Diagnostic accuracy of anti-CN1A on the diagnosis of inclusion body myositis. A hierarchical bivariate and Bayesian meta-analysis. J Clin Neuromuscul Dis. 2021;23:31–8. https://doi.org/10.1097/CND.0000000000000353.
Salam S, Dimachkie MM, Hanna MG, Machado PM. Diagnostic and prognostic value of anti-cN1A antibodies in inclusion body myositis. Clin Exp Rheumatol. 2022;40:384–93. https://doi.org/10.55563/clinexprheumatol/r625rm.
Uruha A, Noguchi S, Hayashi YK, Tsuburaya RS, Yonekawa T, Nonaka I, et al. Hepatitis C virus infection in inclusion body myositis: a case-control study. Neurology. 2016;86:211–7. https://doi.org/10.1212/WNL.0000000000002291.
Wijnbergen D, Johari M, Ozisik O, t Hoen PAC, Ehrhart F, Baudot A, et al. Multi-omics analysis in inclusion body myositis identifies mir-16 responsible for HLA overexpression. Orphanet J Rare Dis. 2025;20:27. https://doi.org/10.1186/s13023-024-03526-x.
Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. https://doi.org/10.1126/science.1134108.
Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–68. https://doi.org/10.1038/nn.2779.
Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–8. https://doi.org/10.1038/nn.2778.
Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349:650–5. https://doi.org/10.1126/science.aab0983.
Britson KA, Ling JP, Braunstein KE, Montagne JM, Kastenschmidt JM, Wilson A, et al. Loss of TDP-43 function and rimmed vacuoles persist after T cell depletion in a xenograft model of sporadic inclusion body myositis. Sci Transl Med. 2022;14:eabi9196. https://doi.org/10.1126/scitranslmed.abi9196.
Nelke C, Kleefeld F, Preusse C, Ruck T, Stenzel W. Inclusion body myositis and associated diseases: an argument for shared immune pathologies. Acta Neuropathol Commun. 2022;10:84. https://doi.org/10.1186/s40478-022-01389-6.
Gang Q, Bettencourt C, Machado P, Hanna MG, Houlden H. Sporadic inclusion body myositis: the genetic contributions to the pathogenesis. Orphanet J Rare Dis. 2014;9:88. https://doi.org/10.1186/1750-1172-9-88.
Weihl CC, Baloh RH, Lee Y, Chou TF, Pittman SK, Lopate G, et al. Targeted sequencing and identification of genetic variants in sporadic inclusion body myositis. Neuromuscul Disord. 2015;25:289–96. https://doi.org/10.1016/j.nmd.2014.12.009.
Gang Q, Bettencourt C, Machado PM, Brady S, Holton JL, Pittman AM, et al. Rare variants in SQSTM1 and VCP genes and risk of sporadic inclusion body myositis. Neurobiol Aging. 2016;47:218.e1–218.e9. https://doi.org/10.1016/j.neurobiolaging.2016.07.024.
Güttsches AK, Brady S, Krause K, Maerkens A, Uszkoreit J, Eisenacher M, et al. Proteomics of rimmed vacuoles define new risk allele in inclusion body myositis. Ann Neurol. 2017;81:227–39. https://doi.org/10.1002/ana.24847.
Hedberg-Oldfors C, Lindgren U, Basu S, Visuttijai K, Lindberg C, Falkenberg M, et al. Mitochondrial DNA variants in inclusion body myositis characterized by deep sequencing. Brain Pathol. 2021;31:e12931. https://doi.org/10.1111/bpa.12931.
Kleefeld F, Uruha A, Schänzer A, Nishimura A, Roos A, Schneider U, et al. Morphologic and molecular patterns of polymyositis with mitochondrial pathology and inclusion body myositis. Neurology. 2022;99:e2212–e2222. https://doi.org/10.1212/WNL.0000000000201103.
Kleefeld F, Cross E, Lagos D, Walli S, Schoser B, Hentschel A, et al. Mitochondrial damage is associated with an early immune response in inclusion body myositis. Brain. 2025:awaf118. https://doi.org/10.1093/brain/awaf118.
Mastaglia FL, Rojana-udomsart A, James I, Needham M, Day TJ, Kiers L, et al. Polymorphism in the TOMM40 gene modifies the risk of developing sporadic inclusion body myositis and the age of onset of symptoms. Neuromuscul Disord. 2013;23:969–74. https://doi.org/10.1016/j.nmd.2013.09.008.
Gang Q, Bettencourt C, Machado PM, Fox Z, Brady S, Healy E, et al. The effects of an intronic polymorphism in TOMM40 and APOE genotypes in sporadic inclusion body myositis. Neurobiol Aging. 2015;36:1766.e1–1766.e3. https://doi.org/10.1016/j.neurobiolaging.2014.12.039.
Nagy S, Khan A, Machado PM, Houlden H. Inclusion body myositis: from genetics to clinical trials. J Neurol. 2023;270:1787–97. https://doi.org/10.1007/s00415-022-11459-3.
Hoogendijk JE, Amato AA, Lecky BR, Choy EH, Lundberg IE, Rose MR, et al. 119th ENMC international workshop: trial design in adult idiopathic inflammatory myopathies, with the exception of inclusion body myositis, 10-12 October 2003, Naarden, The Netherlands. Neuromuscul Disord. 2004;14:337–45. https://doi.org/10.1016/j.nmd.2004.02.006.
Watanabe Y, Uruha A, Suzuki S, Nakahara J, Hamanaka K, Takayama K, et al. Clinical features and prognosis in anti-SRP and anti-HMGCR necrotising myopathy. J Neurol Neurosurg Psychiatry. 2016;87:1038–44. https://doi.org/10.1136/jnnp-2016-313166.
Kurashige T, Nakamura R, Murao T, Mine N, Sato M, Katsumata R, et al. Atypical skin conditions of the neck and back as a dermal manifestation of anti-HMGCR antibody-positive myopathy. BMC Immunol. 2024;25:30 https://doi.org/10.1186/s12865-024-00622-2.
Ohnuki Y, Suzuki S, Uruha A, Oyama M, Suzuki S, Kulski JK, et al. Association of immune-mediated necrotizing myopathy with HLA polymorphisms. HLA. 2023;101:449–57. https://doi.org/10.1111/tan.14950.
Mammen AL, Gaudet D, Brisson D, Christopher-Stine L, Lloyd TE, Leffell MS, et al. Increased frequency of DRB1*11:01 in anti-hydroxymethylglutaryl-coenzyme A reductase-associated autoimmune myopathy. Arthritis Care Res. 2012;64:1233–7. https://doi.org/10.1002/acr.21671.
Liang WC, Uruha A, Suzuki S, Murakami N, Takeshita E, Chen WZ, et al. Pediatric necrotizing myopathy associated with anti-3-hydroxy-3-methylglutaryl-coenzyme A reductase antibodies. Rheumatology. 2017;56:287–93. https://doi.org/10.1093/rheumatology/kew386.
Kishi T, Rider LG, Pak K, Barillas-Arias L, Henrickson M, McCarthy PL, et al. Association of anti-3-hydroxy-3-methylglutaryl-coenzyme a reductase autoantibodies with DRB1*07:01 and severe myositis in juvenile myositis patients. Arthritis Care Res. 2017;69:1088–94. https://doi.org/10.1002/acr.23113.
Ayaki T, Murata K, Kanazawa N, Uruha A, Ohmura K, Sugie K, et al. Myositis with sarcoplasmic inclusions in Nakajo-Nishimura syndrome: a genetic inflammatory myopathy. Neuropathol Appl Neurobiol. 2020;46:579–87.
Jia T, Zheng Y, Feng C, Yang T, Geng S. A Chinese case of Nakajo-Nishimura syndrome with novel compound heterozygous mutations of the PSMB8 gene. BMC Med Genet. 2020;21:126.
Ghodbane NE, Mecibah A, Merzougui Z, Zerguine H, Akakba Z, Slimani S. Nakajo-Nishimura syndrome: the first African Case. Mediterr J Rheumatol. 2023;34:262–5.
Papendorf JJ, Ebstein F, Alehashemi S, Piotto DGP, Kozlova A, Terreri MT, et al. Identification of eight novel proteasome variants in five unrelated cases of proteasome-associated autoinflammatory syndromes (PRAAS). Front Immunol. 2023;14:1190104. https://doi.org/10.3389/fimmu.2023.1190104.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Uruha, A. Genetics of myositis – distinct backgrounds of subtypes. J Hum Genet (2025). https://doi.org/10.1038/s10038-025-01374-6
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s10038-025-01374-6