Abstract
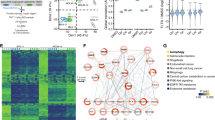
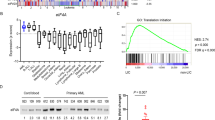
Internal tandem duplication mutations of FLT3 (FLT3/ITD) confer poor prognosis in AML. FLT3 tyrosine kinase inhibitors (TKIs) alone have limited and transient clinical efficacy thus calling for new targets for more effective combination therapy. In a loss-of-function RNAi screen, we identified NOTCH4 as one such potential target whose inhibition proved cytotoxic to AML cells, and also sensitized them to FLT3 inhibition. Further investigation found increased NOTCH4 expression in FLT3/ITD AML cell lines and primary patient samples. Inhibition of NOTCH4 by shRNA knockdown, CRISPR-Cas9-based knockout or γ-secretase inhibitors synergized with FLT3 TKIs to kill FLT3/ITD AML cells in vitro. NOTCH4 inhibition sensitized TKI-resistant FLT3/ITD cells to FLT3 TKI inhibition. The combination reduced phospho-ERK and phospho-AKT, indicating inhibition of MAPK and PI3K/AKT signaling pathways. It also led to changes in expression of genes involved in regulating cell cycling, DNA repair and transcription. A patient-derived xenograft model showed that the combination reduced both the level of leukemic involvement of primary human FLT3/ITD AML cells and their ability to engraft secondary recipients. In summary, these results demonstrate that NOTCH4 inhibition synergizes with FLT3 TKIs to eliminate FLT3/ITD AML cells, providing a new therapeutic target for AML with FLT3/ITD mutations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data generated and analyzed in this study are included in this published article and its supplementary materials and figures.
References
Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–42.
Levis M, Small D. FLT3: ITDoes matter in leukemia. Leukemia. 2003;17:1738–52.
Small D. FLT3 mutations: biology and treatment. Hematology Am Soc Hematol Educ Program. 2006;1:178–84.
Cancer Genome Atlas, Network Research, Ley TJ, Miller C, Ding L, Raphael BJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74.
Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–78.
Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–89.
Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019;33:299–312.
Sutamtewagul G, Vigil CE. Clinical use of FLT3 inhibitors in acute myeloid leukemia. Onco Targets Ther. 2018;11:7041–52.
Perl AE. Availability of FLT3 inhibitors: how do we use them? Blood. 2019;134:741–745.
Chu SH, Small D. Mechanisms of resistance to FLT3 inhibitors. Drug Resist Updat. 2009;12:8–16.
Kayser S, Levis MJ. FLT3 tyrosine kinase inhibitors in acute myeloid leukemia: clinical implications and limitations. Leuk Lymphoma. 2014;55:243–55.
Eguchi M, Minami Y, Kuzume A, Chi S. Mechanisms underlying resistance to FLT3 Inhibitors in acute myeloid leukemia. Biomedicines. 2020;8:245.
Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110.
Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–76.
Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–73.
Vercauteren SM, Sutherland HJ. Constitutively active Notch4 promotes early human hematopoietic progenitor cell maintenance while inhibiting differentiation and causes lymphoid abnormalities in vivo. Blood. 2004;104:2315–22.
Ye Q, Shieh JH, Morrone G, Moore MA. Expression of constitutively active Notch4 (Int-3) modulates myeloid proliferation and differentiation and promotes expansion of hematopoietic progenitors. Leukemia. 2004;18:777–87.
Li Y, Jin C, Bai H, Gao Y, Sun S, Chen L, et al. Human NOTCH4 is a key target of RUNX1 in megakaryocytic differentiation. Blood. 2018;131:191–201.
Kannan S, Sutphin RM, Hall MG, Golfman LS, Fang W, Nolo RM, et al. Notch activation inhibits AML growth and survival: a potential therapeutic approach. J Exp Med. 2013;210:321–37.
Xiu M, Zeng X, Shan R, Wen W, Li J, Wan R. Targeting Notch4 in cancer: Molecular mechanisms and therapeutic perspectives. Cancer Manag Res. 2021;13:7033–45.
Piloto O, Wright M, Brown P, Kim KT, Levis M, Small D. Prolonged exposure to FLT3 inhibitors leads to resistance via activation of parallel signaling pathways. Blood. 2007;109:1643–52.
Zhu R, Li L, Nguyen B, Seo J, Wu M, Seale T, et al. FLT3 tyrosine kinase inhibitors synergize with BCL-2 inhibition to eliminate FLT3/ITD acute leukemia cells through BIM activation. Signal Transduct Target Ther. 2021;6:186.
Huang A, Ju HQ, Liu K, Zhan G, Liu D, Wen S, et al. Metabolic alterations and drug sensitivity of tyrosine kinase inhibitor resistant leukemia cells with a FLT3/ITD mutation. Cancer let. 2016;377:149–57.
Stirewalt DL, Meshinchi S, Kopecky KJ, Fan W, Pogosova-Agadjanyan EL, Engel JH, et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosomes Cancer. 2008;47:8–20.
Li L, Piloto O, Nguyen HB, Greenberg K, Takamiya K, Racke F, et al. Knock-in of an internal tandem duplication mutation into murine FLT3 confers myeloproliferative disease in a mouse model. Blood. 2008;111:3849–58.
Li L, Bailey E, Greenblatt S, Huso D, Small D. Loss of the wild-type allele contributes to myeloid expansion and disease aggressiveness in FLT3/ITD knockin mice. Blood. 2011;118:4935–45.
Bailey E, Li L, Duffield AS, Ma HS, Huso DL, Small D. FLT3/D835Y mutation knock-in mice display less aggressive disease compared with FLT3/internal tandem duplication (ITD) mice. Proc Natl Acad Sci. 2013;110:21113–8.
Tarumoto Y, Lu B, Somerville TDD, Huang YH, Milazzo JP, Wu XS, et al. LKB1, Salt-Inducible Kinases, and MEF2C are linked dependencies in acute myeloid leukemia. Mol Cell. 2018;69:1017–27.
Knudsen ES, Witkiewicz AK, Rubin SM. Cancer takes many paths through G1/S. Trends Cell Biol. 2023;S0962–8924:00211–8.
Oh P, Lobry C, Gao J, Tikhonova A, Loizou E, Manet J, et al. In vivo mapping of notch pathway activity in normal and stress hematopoiesis. Cell Stem Cell. 2013;13:190–204.
Mercher T, Cornejo MG, Sears C, Kindler T, Moore SA, Maillard I, et al. Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell Stem Cell. 2008;3:314–26.
Takam Kamga P, Dal Collo G, Resci F, Bazzoni R, Mercuri A, Quaglia FM, et al. Notch signaling molecules as prognostic biomarkers for acute myeloid leukemia. Cancers. 2019;11:1958.
Weng AP, Ferrando AA, Lee W, Morris JP 4th, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71.
Lobry C, Oh P, Mansour MR, Look AT, Aifantis I. Notch signaling: switching an oncogene to a tumor suppressor. Blood. 2014;123:2451–59.
Arcaini L, Rossi D, Lucioni M, Nicola M, Bruscaggin A, Fiaccadori V, et al. The NOTCH pathway is recurrently mutated in diffuse large B-cell lymphoma associated with hepatitis C virus infection. Haematologica. 2015;100:246–52.
Kridel R, Meissner B, Rogic S, Boyle M, Telenius A, Woolcock B, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119:63–1971.
Rossi D, Trifonov V, Fangazio M, Bruscaggin A, Rasi S, Spina V, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209:1537–51.
Koch U, Radtke F. Notch signaling in solid tumors. Curr Top Dev Biol. 2010;92:411–55.
Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–51.
Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N, et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J Exp Med. 2013;210:301–19.
Tohda S, Sakano S, Ohsawa M, Murakami N, Nara N. A novel cell line derived from de novo acute myeloblastic leukaemia with trilineage myelodysplasia which proliferates in response to a Notch ligand, Delta-1 protein. Br J Haematol. 2002;117:373–78.
Kode A, Manavalan JS, Mosialou I, Bhagat G, Rathinam CV, Luo N, et al. Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature. 2014;506:240–44.
Li D, Li T, Shang Z, Zhao L, Xu Q, Tan J, et al. Combined inhibition of Notch and FLT3 produces synergistic cytotoxic effects in FLT3/ITD(+) acute myeloid leukemia. Signal Transduct Target Ther. 2020;5:21.
Le Q, Hadland B, Meshinchi S, Bernstein I. Notch blockade overcomes endothelial cell-mediated resistance of FLT3/ITD-positive AML progenitors to AC220 treatment. Leukemia. 2021;35:601–05.
Bazarbachi AH, Al Hamed R, Malard F, Mohty M, Bazarbachi A. Allogeneic transplant for FLT3-ITD mutated AML: a focus on FLT3 inhibitors before, during, and after transplant. Ther Adv Hematol. 2019;10:2040620719882666.
Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–63.
Sato T, Yang X, Knapper S, White P, Smith BD, Galkin S, et al. FLT3 ligand impedes the efficacy of FLT3 inhibitors in vitro and in vivo. Blood. 2011;117:3286–93.
Yang X, Sexauer A, Levis M. Bone marrow stroma-mediated resistance to FLT3 inhibitors in FLT3-ITD AML is mediated by persistent activation of extracellular regulated kinase. Br J Haematol. 2014;164:61–72.
Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86.
Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71.
Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell. 2012;22:668–82.
Bruner JK, Ma HS, Li L, Qin ACR, Rudek MA, Jones RJ, et al. Adaptation to TKI Treatment Reactivates ERK Signaling in Tyrosine Kinase-Driven Leukemias and Other Malignancies. Cancer Res. 2017;77:5554–63.
Acknowledgements
This work was supported by grants from the National Cancer Institute (R01 CA090668 (DS) and P30 CA006973, T32 CA60441 (CMS), Commonwealth Fund and the Giant Food Pediatric Cancer Research Fund (DS). RZ is supported by the China Scholarship Council and National Natural Science Foundation of China (No. 82200187). DS is also supported by the Kyle Haydock Professorship. DS has also received research funding and has served as a consultant to Pharos I, B&T for an unrelated project and serves on the SAB of In Silico Medicine, Inc. and Mitomed.
Author information
Authors and Affiliations
Contributions
RZ, CMS and SHC designed experiments, performed experiments and analyzed data; LL, BHN, JS, MW and AD performed data analysis and provided technical support; LMS and YH provided technical support; ML provided patient samples; DS designed experiments and analyzed data. RZ wrote the first version of the manuscript; RZ, CMS, LL and DS revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interest. DS does serve on the SAB of InSilico Medicine and Mitomed and receives research support and serves as a consultant for an unrelated project from Pharos, Ltd. ML has received honoraria from Daiichi Sankyo, Novartis, and Agios; has served in a consulting or advisory role for Daiichi Sankyo, Novartis, and Agios; and has received research funding from Astellas and Novartis.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhu, R., Shirley, C.M., Chu, S.H. et al. Inhibition of NOTCH4 sensitizes FLT3/ITD acute myeloid leukemia cells to FLT3 tyrosine kinase inhibition. Leukemia 38, 1581–1591 (2024). https://doi.org/10.1038/s41375-024-02292-w
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41375-024-02292-w
This article is cited by
-
Targeting leukemic stem cell biomechanics suppresses stemness and enhances NK cell-mediated immunotherapy
Nature Communications (2025)