Abstract
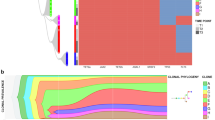
Myeloproliferative neoplasms (MPNs) are characterized by clonal proliferation of hematopoietic stem cells, which can lead to secondary myelofibrosis or acute myeloid leukemia. We explored the changes in genomic alterations during MPN transformation using whole-genome sequencing of samples from both the chronic and fibrotic or leukemic phases of 20 patients. We identified FOXP1 mutations in 3 of 14 (21.4%) patients with secondary myelofibrosis. This novel mutation was identified in another 5 of the 35 patients (14.3%) in an independent cohort. All these 8 patients with FOXP1 mutations did not experience leukemic transformation after a median follow-up of 5.1 years. The acquisition of non-canonical MPLY591 mutations was detected in the fibrotic or leukemic phase. Clonal expansion, involving both known and unknown driver genes (in 18 and 2 patients, respectively), was observed in all patients. We determined the patterns of clonal evolution based on myeloid driver mutations in 18 patients: linear clonal evolution in 11 patients and branched clonal evolution in 7 patients. Our results suggested that MPN patients carrying FOXP1 mutations are unlikely to have leukemia transformation and emphasized that the acquisition of specific genetic mutations and dynamic changes in clonal architecture underlie the pathogenesis in patients undergoing MPN transformation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated during and analyzed during the current study are available from the corresponding authors on reasonable request. CNA data are available at the following link: https://github.com/HiroyukiTakamori/WGS_MPN_Progression.
References
Thiele J, Kvasnicka HM, Orazi A, Gianelli U, Gangat N, Vannucchi AM, et al. The international consensus classification of myeloid neoplasms and acute Leukemias: myeloproliferative neoplasms. Am J Hematol. 2023;98:166–79.
Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka H-M, et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140:1200–28.
Tefferi A, Alkhateeb H, Gangat N. Blast phase myeloproliferative neoplasm: contemporary review and 2024 treatment algorithm. Blood Cancer J. 2023;13. 108.
Cerquozzi S, Tefferi A. Blast transformation and fibrotic progression in polycythemia vera and essential thrombocythemia: a literature review of incidence and risk factors. Blood Cancer J. 2015;5.e366.
Szuber N, Mudireddy M, Nicolosi M, Penna D, Vallapureddy RR, Lasho TL, et al. 3023 Mayo Clinic Patients With Myeloproliferative Neoplasms: Risk-Stratified Comparison of Survival and Outcomes Data Among Disease Subgroups. Mayo Clin Proc. 2019;94:599–610.
Tam CS, Kantarjian H, Cortes J, Lynn A, Pierce S, Zhou L, et al. Dynamic model for predicting death within 12 months in patients with primary or post-polycythemia vera/essential thrombocythemia myelofibrosis. J Clin Oncol. 2009;27:5587–93.
Mudireddy M, Gangat N, Hanson CA, Ketterling RP, Pardanani A, Tefferi A. Validation of the WHO-defined 20% circulating blasts threshold for diagnosis of leukemic transformation in primary myelofibrosis. Blood Cancer J. 2018;8:57.
Tefferi A, Mudireddy M, Mannelli F, Begna KH, Patnaik MM, Hanson CA, et al. Blast phase myeloproliferative neoplasm: Mayo-AGIMM study of 410 patients from two separate cohorts. Leukemia. 2018;32:1200–10.
Kennedy JA, Atenafu EG, Messner HA, Craddock KJ, Brandwein JM, Lipton JH, et al. Treatment outcomes following leukemic transformation in Philadelphia-negative myeloproliferative neoplasms. Blood. 2013;121:2725–33.
Patel AA, Yoon JJ, Johnston H, Davidson MB, Shallis RM, Chen EC, et al. Treatment approach and outcomes of patients with accelerated/blast-phase myeloproliferative neoplasms in the current era. Blood Adv. 2024. https://doi.org/10.1182/bloodadvances.2024012880.
Grinfeld J, Nangalia J, Baxter EJ, Wedge DC, Angelopoulos N, Cantrill R, et al. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. N Engl J Med. 2018;379:1416–30.
Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I, et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123:2220–8.
Vannucchi AM, Lasho TL, Guglielmelli P, Biamonte F, Pardanani A, Pereira A, et al. Mutations and prognosis in primary myelofibrosis. Leukemia. 2013;27:1861–9.
Tefferi A, Lasho TL, Finke CM, Elala Y, Hanson CA, Ketterling RP, et al. Targeted deep sequencing in primary myelofibrosis. Blood Adv. 2016;1:105–11.
Tefferi A, Vannucchi AM. Genetic Risk Assessment in Myeloproliferative Neoplasms. Mayo Clin Proc. 2017;92:1283–90.
Tefferi A, Lasho TL, Guglielmelli P, Finke CM, Rotunno G, Elala Y, et al. Targeted deep sequencing in polycythemia vera and essential thrombocythemia. Blood Adv. 2016;1:21–30.
Nangalia J, Campbell PJ. Genome Sequencing during a Patient’s Journey through Cancer. N Engl J Med. 2019;381:2145–56.
Duncavage EJ, Schroeder MC, O’Laughlin M, Wilson R, MacMillan S, Bohannon A, et al. Genome Sequencing as an Alternative to Cytogenetic Analysis in Myeloid Cancers. N Engl J Med. 2021;384:924–35.
Engle EK, Fisher DAC, Miller CA, McLellan MD, Fulton RS, Moore DM, et al. Clonal evolution revealed by whole genome sequencing in a case of primary myelofibrosis transformed to secondary acute myeloid leukemia. Leukemia. 2015;29:869–76.
Williams N, Lee J, Mitchell E, Moore L, Baxter EJ, Hewinson J, et al. Life histories of myeloproliferative neoplasms inferred from phylogenies. Nature. 2022;602:162–8.
Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia. 2022;36:1703–19.
Maslah N, Benajiba L, Giraudier S, Kiladjian J-J, Cassinat B. Clonal architecture evolution in Myeloproliferative Neoplasms: from a driver mutation to a complex heterogeneous mutational and phenotypic landscape. Leukemia. 2023;37:957–63.
Guess T, Potts CR, Bhat P, Cartailler JA, Brooks A, Holt C, et al. Distinct Patterns of Clonal Evolution Drive Myelodysplastic Syndrome Progression to Secondary Acute Myeloid Leukemia. Blood Cancer Discov. 2022;3:316–29.
Mitchell J, Milite S, Bartram J, Walker S, Volkova N, Yavorska O, et al. Clinical application of tumour-in-normal contamination assessment from whole genome sequencing. Nat Commun. 2024;15:323.
Tenedini E, Bernardis I, Artusi V, Artuso L, Roncaglia E, Guglielmelli P, et al. Targeted cancer exome sequencing reveals recurrent mutations in myeloproliferative neoplasms. Leukemia. 2014;28:1052–9.
Mylonas E, Yoshida K, Frick M, Hoyer K, Christen F, Kaeda J, et al. Single-cell analysis based dissection of clonality in myelofibrosis. Nat Commun. 2020;11:73.
Marcault C, Zhao L-P, Maslah N, Verger E, Daltro de Oliveira R, Soret-Dulphy J, et al. Impact of NFE2 mutations on AML transformation and overall survival in patients with myeloproliferative neoplasms. Blood. 2021;138:2142–8.
Hung HL, Kim AY, Hong W, Rakowski C, Blobel GA. Stimulation of NF-E2 DNA binding by CREB-binding protein (CBP)-mediated acetylation. J Biol Chem. 2001;276:10715–21.
Jutzi JS, Bogeska R, Nikoloski G, Schmid CA, Seeger TS, Stegelmann F, et al. MPN patients harbor recurrent truncating mutations in transcription factor NF-E2. J Exp Med. 2013;210:1003–19.
Cabagnols X, Favale F, Pasquier F, Messaoudi K, Defour JP, Ianotto JC, et al. Presence of atypical thrombopoietin receptor (MPL) mutations in triple-negative essential thrombocythemia patients. Blood. 2016;127:333–42.
Milosevic Feenstra JD, Nivarthi H, Gisslinger H, Leroy E, Rumi E, Chachoua I, et al. Whole-exome sequencing identifies novel MPL and JAK2 mutations in triple-negative myeloproliferative neoplasms. Blood. 2016;127:325–32.
Hu H, Wang B, Borde M, Nardone J, Maika S, Allred L, et al. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol. 2006;7:819–26.
Shi C, Sakuma M, Mooroka T, Liscoe A, Gao H, Croce KJ, et al. Down-regulation of the forkhead transcription factor Foxp1 is required for monocyte differentiation and macrophage function. Blood. 2008;112:4699–711.
Zhang Y, Li S, Yuan L, Tian Y, Weidenfeld J, Yang J, et al. Foxp1 coordinates cardiomyocyte proliferation through both cell-autonomous and nonautonomous mechanisms. Genes Dev. 2010;24:1746–57.
Li S, Wang Y, Zhang Y, Lu MM, DeMayo FJ, Dekker JD, et al. Foxp1/4 control epithelial cell fate during lung development and regeneration through regulation of anterior gradient 2. Development. 2012;139:2500–9.
Brown PJ, Wong KK, Felce SL, Lyne L, Spearman H, Soilleux EJ, et al. FOXP1 suppresses immune response signatures and MHC class II expression in activated B-cell-like diffuse large B-cell lymphomas. Leukemia. 2016;30:605–16.
Ackermann S, Kocak H, Hero B, Ehemann V, Kahlert Y, Oberthuer A, et al. FOXP1inhibits cell growth and attenuates tumorigenicity of neuroblastoma. BMC Cancer. 2014;14:840.
Takayama K-I, Suzuki T, Tsutsumi S, Fujimura T, Takahashi S, Homma Y, et al. Integrative analysis of FOXP1 function reveals a tumor-suppressive effect in prostate cancer. Mol Endocrinol. 2014;28:2012–24.
Klampfl T, Harutyunyan A, Berg T, Gisslinger B, Schalling M, Bagienski K, et al. Genome integrity of myeloproliferative neoplasms in chronic phase and during disease progression. Blood. 2011;118:167–76.
Shao X, Wei X. FOXP1 enhances fibrosis via activating Wnt/β-catenin signaling pathway in endometriosis. Am J Transl Res. 2018;10:3610–8.
Liu J, Zhuang T, Pi J, Chen X, Zhang Q, Li Y, et al. Endothelial Forkhead Box Transcription Factor P1 Regulates Pathological Cardiac Remodeling Through Transforming Growth Factor-β1-Endothelin-1 Signal Pathway. Circulation. 2019;140:665–80.
Beer PA, Delhommeau F, LeCouédic J-P, Dawson MA, Chen E, Bareford D, et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood. 2010;115:2891–2900.
Knudsen TA, Skov V, Stevenson K, Werner L, Duke W, Laurore C, et al. Genomic profiling of a randomized trial of interferon-α vs hydroxyurea in MPN reveals mutation-specific responses. Blood Adv. 2022;6:2107–19.
Pérez C, Pascual M, Martín-Subero JI, Bellosillo B, Segura V, Delabesse E, et al. Aberrant DNA methylation profile of chronic and transformed classic Philadelphia-negative myeloproliferative neoplasms. Haematologica. 2013;98:1414–20.
Harutyunyan A, Klampfl T, Cazzola M, Kralovics R. P53 lesions in leukemic transformation. N Engl J Med. 2011;364:488–90.
Zhang S-J, Rampal R, Manshouri T, Patel J, Mensah N, Kayserian A, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119:4480–5.
Luque Paz D, Jouanneau-Courville R, Riou J, Ianotto J-C, Boyer F, Chauveau A, et al. Leukemic evolution of polycythemia vera and essential thrombocythemia: genomic profiles predict time to transformation. Blood Adv. 2020;4:4887–97.
Venton G, Courtier F, Charbonnier A, D’incan E, Saillard C, Mohty B, et al. Impact of gene mutations on treatment response and prognosis of acute myeloid leukemia secondary to myeloproliferative neoplasms. Am J Hematol. 2018;93:330–8.
Acknowledgements
The authors acknowledge the patients who participated in the study and their families. The authors thank the clinical research staff and caregivers at all the participating sites. Supercomputing resources were provided by the Human Genome Center, the Institute of Medical Science, the University of Tokyo, Japan.
Funding
This work was supported by grants from the National Science and Technology Council, Taiwan (L-YS: MOST110-2314-B-182-018, MOST111-2314-B-182-043, and NSTC 112-2314-B-182-055), the Ministry of Health and Welfare, Taiwan (L-YS: DOH102-TD-C-111-006), Grant-in-Aid for JSPS KAKENHI (SO: JP19H05656), and the Japan Agency for Medical Research and Development, AMED (YN: 23ama221506h0002).
Author information
Authors and Affiliations
Contributions
L-YS and YN designed and supervised the study. L-YS and Y-JH provided whole- genome sequencing raw data and targeted next generation sequencing data, L-YS and M-CK treated the patients and provided the samples, L-YS, M-CK, and T-YH collected and organized all clinical data. HT, HF, KY, SO, and YN analyzed WGS data. HT, YN, Y-JH and L-YS wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
SO reports a leadership position and advisory role with Eisai Co., Ltd and Chordia Therapeutics, Inc. SO is also a stockholder in Asahi Genomics Co., Ltd., and has received honoraria from The Mitsubishi Foundation and the Nakatani Foundation. Additionally, SO reports receiving grant/research funding from Chordia Therapeutics, Inc., Otsuka Pharmaceutical Co., Ltd., Eisai Co., Ltd, and Nippon Shinyaku Co., Ltd. YN has received grant/research funding from Daiichi Sankyo Co., Ltd., ThinkCyte Co., Ltd., and Otsuka Pharmaceutical Co., Ltd. YN has also received honoraria from Bristol Myers Squibb Co., Ltd., Takeda Pharmaceutical Co., Ltd., Daiichi Sankyo Co., Ltd., Kyowa Hakko Kirin Co., Ltd., Novartis Pharma Co., Ltd., Pfizer Japan Co., Ltd., Nippon Shinyaku Co., Ltd., Otsuka Pharmaceutical Co., Ltd., and AstraZeneca Co., Ltd. The remaining authors declare no competing financial interests related to this study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Takamori, H., Huang, YJ., Fukushima, H. et al. Whole-genome sequencing of myeloproliferative neoplasms revealed dynamic clonal changes in the fibrotic or leukemic transformation and novel FOXP1 mutations in the fibrotic transformation. Leukemia 39, 1218–1227 (2025). https://doi.org/10.1038/s41375-025-02576-9
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41375-025-02576-9