Abstract
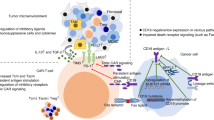
The modest reduction in casein kinase 1 alpha (CK1α) by lenalidomide contributes to its clinical effectiveness in treating del(5q) myelodysplastic syndrome. However, the mechanism by which CK1α impacts lymphoma survival remains inadequately defined. We developed INNO-220, a CRBN-dependent CK1α degrader, by leveraging cytokine expression profiling in T cells. Unlike lenalidomide, INNO-220 is a highly selective and potent degrader of CK1α without affecting IKZF1/3. Screening across lymphoma cell lines revealed that cells harboring wild-type p53 and exhibiting constitutive NF-κB signaling were particularly sensitive to CK1α degradation yet resistant to Bruton tyrosine kinase inhibitors. Moreover, INNO-220 suppresses NF-κB signaling and activates p53 pathway, leading to complete inhibition of lymphoma tumor growth in vivo. Mechanistically, INNO-220 disrupts the assembly and function of the CARD11/BCL10/MALT1 complex, thereby inhibiting NF-κB signaling in stimulated T cells and lymphoma cells that harbor an activating mutation in CARD11. Moreover, we observed that activation of wild-type p53 upon INNO-220 treatment was sufficient to induce potent cancer cell death even in the absence of constitutive NF-κB activity. In summary, our findings introduce a selective CK1α degrader as a novel therapeutic approach for lymphoma, providing both mechanistic insights and a potential patient selection strategy in treating lymphoma and possibly other cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA-seq data are publicly available in Gene Expression Omnibus (GEO) at GSE270521. The data supporting the findings of this study are available from the corresponding author upon reasonable request. All other data are available in the article and its supplementary materials.
References
Silkenstedt E, Salles G, Campo E, Dreyling M. B-cell non-Hodgkin lymphomas. Lancet. 2024;403:1791–807.
Stephens DM, Byrd JC. Resistance to Bruton tyrosine kinase inhibitors: the Achilles heel of their success story in lymphoid malignancies. Blood. 2021;138:1099–109.
Zhong G, Kong R, Feng S, Wang C, Hao Q, Xie W, et al. Targeted protein degradation in hematologic malignancies: latest updates from the 2023 ASH annual meeting. J Hematol Oncol. 2024;17:14.
Jan M, Sperling AS, Ebert BL. Cancer therapies based on targeted protein degradation - lessons learned with lenalidomide. Nat Rev Clin Oncol. 2021;18:401–17.
Kozicka Z, Thoma NH. Haven’t got a glue: Protein surface variation for the design of molecular glue degraders. Cell Chem Biol. 2021;28:1032–47.
Goldhirsh G, Kravtsova-Ivantsiv Y, Satish G, Ziv T, Brik A, Ciechanover A. A short binding site in the KPC1 ubiquitin ligase mediates processing of NF-kappaB1 p105 to p50: a potential for a tumor-suppressive PROTAC. Proc Natl Acad Sci USA. 2021;118:e2117254118.
Zhong G, Chang X, Xie W, Zhou X. Targeted protein degradation: advances in drug discovery and clinical practice. Signal Transduct Target Ther. 2024;9:308.
Gandhi AK, Kang J, Havens CG, Conklin T, Ning Y, Wu L, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN. Br J Haematol. 2014;164:811–21.
Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–5.
Drula R, Iluta S, Gulei D, Iuga C, Dima D, Ghiaur G, et al. Exploiting the ubiquitin system in myeloid malignancies. From basic research to drug discovery in MDS and AML. Blood Rev. 2022;56:100971.
Chamberlain PP, Lopez-Girona A, Miller K, Carmel G, Pagarigan B, Chie-Leon B, et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat Struct Mol Biol. 2014;21:803–9.
Kronke J, Fink EC, Hollenbach PW, MacBeth KJ, Hurst SN, Udeshi ND, et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature. 2015;523:183–8.
Petzold G, Fischer ES, Thoma NH. Structural basis of lenalidomide-induced CK1alpha degradation by the CRL4(CRBN) ubiquitin ligase. Nature. 2016;532:127–30.
Groppe JC. Induced degradation of protein kinases by bifunctional small molecules: a next-generation strategy. Expert Opin Drug Discov. 2019;14:1237–53.
Roskoski R Jr. A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol Res. 2015;100:1–23.
Bartlett JB, Dredge K, Dalgleish AG. The evolution of thalidomide and its IMiD derivatives as anticancer agents. Nat Rev Cancer. 2004;4:314–22.
Martinez-Valbuena I, Lee S, Santamaria E, Irigoyen JF, Forrest S, Li J, et al. 4R-Tau seeding activity unravels molecular subtypes in patients with progressive supranuclear Palsy. bioRxiv. 2023.
Zhou X, Chen N, Xu H, Zhou X, Wang J, Fang X, et al. Regulation of Hippo-YAP signaling by insulin-like growth factor-1 receptor in the tumorigenesis of diffuse large B-cell lymphoma. J Hematol Oncol. 2020;13:77.
Chen X, Lu T, Cai Y, Han Y, Ding M, Chu Y, et al. KIAA1429-mediated m6A modification of CHST11 promotes progression of diffuse large B-cell lymphoma by regulating Hippo-YAP pathway. Cell Mol Biol Lett. 2023;28:32.
Huart AS, MacLaine NJ, Meek DW, Hupp TR. CK1alpha plays a central role in mediating MDM2 control of p53 and E2F-1 protein stability. J Biol Chem. 2009;284:32384–94.
Chen L, Li C, Pan Y, Chen J. Regulation of p53-MDMX interaction by casein kinase 1 alpha. Mol Cell Biol. 2005;25:6509–20.
Järås M, Miller PG, Chu LP, Puram RV, Fink EC, Schneider RK, et al. Csnk1a1 inhibition has p53-dependent therapeutic efficacy in acute myeloid leukemia. J Exp Med. 2014;211:605–12.
Ruefli-Brasse AA, French DM, Dixit VM. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science. 2003;302:1581–4.
Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 2006;25:701–15.
Bidere N, Ngo VN, Lee J, Collins C, Zheng L, Wan F, et al. Casein kinase 1alpha governs antigen-receptor-induced NF-kappaB activation and human lymphoma cell survival. Nature. 2009;458:92–96.
Häcker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006:re13.
Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, et al. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–4.
Gehring T, Erdmann T, Rahm M, Grass C, Flatley A, O’Neill TJ, et al. MALT1 phosphorylation controls activation of T lymphocytes and survival of ABC-DLBCL tumor cells. Cell Rep. 2019;29:873–88.e810.
Carvalho G, Le Guelte A, Demian C, Vazquez A, Gavard J, Bidère N. Interplay between BCL10, MALT1 and IkappaBalpha during T-cell-receptor-mediated NFkappaB activation. J Cell Sci. 2010;123:2375–80.
Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2:a000109.
Knies N, Alankus B, Weilemann A, Tzankov A, Brunner K, Ruff T, et al. Lymphomagenic CARD11/BCL10/MALT1 signaling drives malignant B-cell proliferation via cooperative NF-kappaB and JNK activation. Proc Natl Acad Sci USA. 2015;112:E7230–38.
Smith CIE, Burger JA. Resistance mutations to BTK inhibitors originate from the NF-kappaB but not from the PI3K-RAS-MAPK arm of the B cell receptor signaling pathway. Front Immunol. 2021;12:689472.
Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–9.
Tisato V, Voltan R, Gonelli A, Secchiero P, Zauli G. MDM2/X inhibitors under clinical evaluation: perspectives for the management of hematological malignancies and pediatric cancer. J Hematol Oncol. 2017;10:133.
Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299–309.
Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci USA. 2003;100:12009–14.
Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–57.
Leslie PL, Ke H, Zhang Y. The MDM2 RING domain and central acidic domain play distinct roles in MDM2 protein homodimerization and MDM2-MDMX protein heterodimerization. J Biol Chem. 2015;290:12941–50.
Wu S, Chen L, Becker A, Schonbrunn E, Chen J. Casein kinase 1alpha regulates an MDMX intramolecular interaction to stimulate p53 binding. Mol Cell Biol. 2012;32:4821–32.
Ruland J, Hartjes L. CARD-BCL-10-MALT1 signalling in protective and pathological immunity. Nat Rev Immunol. 2019;19:118–34.
Matsumoto R, Wang D, Blonska M, Li H, Kobayashi M, Pappu B, et al. Phosphorylation of CARMA1 plays a critical role in T Cell receptor-mediated NF-kappaB activation. Immunity. 2005;23:575–85.
Sommer K, Guo B, Pomerantz JL, Bandaranayake AD, Moreno-Garcia ME, Ovechkina YL, et al. Phosphorylation of the CARMA1 linker controls NF-kappaB activation. Immunity. 2005;23:561–74.
Manni S, Fregnani A, Quotti Tubi L, Spinello Z, Carraro M, Scapinello G, et al. Protein Kinase CK1alpha Sustains B-Cell Receptor Signaling in Mantle Cell Lymphoma. Front Oncol. 2021;11:733848.
Cheng J, Hamilton KS, Kane LP. Phosphorylation of Carma1, but not Bcl10, by Akt regulates TCR/CD28-mediated NF-kappaB induction and cytokine production. Mol Immunol. 2014;59:110–6.
Schittek B, Sinnberg T. Biological functions of casein kinase 1 isoforms and putative roles in tumorigenesis. Mol Cancer. 2014;13:231.
Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10.
Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96.
Karni-Schmidt O, Lokshin M, Prives C. The Roles of MDM2 and MDMX in Cancer. Annu Rev Pathol. 2016;11:617–44.
Fang Y, Zhang MC, He Y, Li C, Fang H, Xu PP, et al. Human endogenous retroviruses as epigenetic therapeutic targets in TP53-mutated diffuse large B-cell lymphoma. Signal Transduct Target Ther. 2023;8:381.
Lew TE, Minson A, Dickinson M, Handunnetti SM, Blombery P, Khot A, et al. Treatment approaches for patients with TP53-mutated mantle cell lymphoma. Lancet Haematol. 2023;10:e142–e154.
Minzel W, Venkatachalam A, Fink A, Hung E, Brachya G, Burstain I, et al. Small molecules co-targeting CKIalpha and the transcriptional kinases CDK7/9 control AML in preclinical models. Cell. 2018;175:171–85.e125.
Nishiguchi G, Mascibroda LG, Young SM, Caine EA, Abdelhamed S, Kooijman JJ, et al. Selective CK1alpha degraders exert antiproliferative activity against a broad range of human cancer cell lines. Nat Commun. 2024;15:482.
Park SM, Miyamoto DK, Han GYQ, Chan M, Curnutt NM, Tran NL, et al. Dual IKZF2 and CK1alpha degrader targets acute myeloid leukemia cells. Cancer Cell. 2023;41:726–39.e711.
Kanaoka D, Yamada M, Yokoyama H, Nishino S, Kunimura N, Satoyoshi H, et al. FPFT-2216, a novel anti-lymphoma compound, induces simultaneous degradation of IKZF1/3 and CK1alpha to activate p53 and inhibit NFkappaB signaling. Cancer Res Commun. 2024;4:312–27.
Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92.
Phelan JD, Young RM, Webster DE, Roulland S, Wright GW, Kasbekar M, et al. A multiprotein supercomplex controlling oncogenic signalling in lymphoma. Nature. 2018;560:387–91.
Young RM, Shaffer AL 3rd, Phelan JD, Staudt LM. B-cell receptor signaling in diffuse large B-cell lymphoma. Semin Hematol. 2015;52:77–85.
Nalawansha DA, Crews CM. PROTACs: an emerging therapeutic modality in precision medicine. Cell Chem Biol. 2020;27:998–1014.
Li K, Crews CM. PROTACs: past, present and future. Chem Soc Rev. 2022;51:5214–36.
Funding
National Natural Science Foundation (No. 82170189). Taishan Scholars Program of Shandong Province. Shandong Provincial Natural Science Foundation (ZR2021YQ51). Technology Development Project of Jinan City (No. 202134034).
Author information
Authors and Affiliations
Contributions
W.X. and X.Z. designed research; S.F., R.K., C.W., Q.H., X.X., Y.Z., J.E., D.M., M.H., V.P.K., P.P. and F.M. performed research; X.Z., W.X., S.F. analyzed data; J.H. and P.K. contributed new reagents/analytic tools; and S.F., R.K., C.W., Q.H., F.M., W.X. and X.Z. wrote the paper. S.F., R.K., Q.H. and C.W. contributed equally to this work.
Corresponding authors
Ethics declarations
Competing interests
W.X., J.E., D.M., M.H., P.K., V.P.K., P.P., and F.M. are currently employed by Innovo Therapeutics and currently hold stock options in a privately held company. This study did not receive funding from Innovo Therapeutics, and we declare that these relationships do not influence the study design, data analysis, or interpretation of results. The remaining authors declare that they have no competing interests.
Ethics approval
This study was approved by the Medical Ethical Committee of Shandong Provincial Hospital. All samples were obtained with informed consent in accordance with the Declaration of Helsinki. All animal experimental procedures were performed in accordance with the protocols approved by the Institutional Animal Care and Research Advisory Committee of Shandong Provincial Hospital.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Feng, S., Kong, R., Wang, C. et al. A highly selective and orally bioavailable casein kinase 1 alpha degrader through p53 signaling pathway targets B-cell lymphoma cells. Leukemia 39, 1972–1986 (2025). https://doi.org/10.1038/s41375-025-02647-x
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41375-025-02647-x