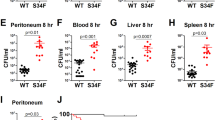
Abstract
U2AF1 mutations are common in patients with myelodysplastic neoplasms (MDS), suggesting that aberrant splicing of pre-mRNAs driven by mutant U2AF1 could play a critical role in MDS pathogenesis. Previous studies have demonstrated that U2AF1S34F mutation impairs the differentiation of erythrocytes and granulocytes, but the impact on megakaryocytes (MKs) remains unclear. Here, by integrating data from MDS patients and cell lines with U2AF1 mutations, we determined that U2AF1 mutations are associated with dysmegakaryopoiesis, induce the generation of abnormal MKs, especially micro-MKs, and induce significant thrombocytopenia. We determined that mutant U2AF1-mediated aberrant splicing of DNA biosynthesis-related genes, such as CHEK1, is required for normal MK polyploidization. The mis-splicing of CHEK1, in turn, accounts for the increased number of abnormal MKs in U2AF1-mutant MDS patients. Moreover, U2AF1S34 mutations induce the deficiency of CHK1 and the activation of its phosphorylation, thereby further driving the impairment of MK polyploidization and maturation. Accordingly, treatment with selective CHK1 inhibitor significantly reduces abnormal MK production in vitro. Taken together, these findings demonstrate that U2AF1 mutations induce the generation of abnormal MKs by driving aberrant splicing of the CHEK1 cell cycle-related gene, revealing the molecular basis for dysmegakaryopoiesis in MDS and identifying a new potential target for MDS treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets supporting the findings of this study are available from the corresponding authors upon reasonable request.
References
Cazzola M. Myelodysplastic syndromes. N Engl J Med. 2020;383:1358–74.
Ogawa S. Genetics of MDS. Blood. 2019;133:1049–59.
Fabre MA, de Almeida JG, Fiorillo E, Mitchell E, Damaskou A, Rak J, et al. The longitudinal dynamics and natural history of clonal haematopoiesis. Nature. 2022;606:335–42.
van Zeventer IA, de Graaf AO, Salzbrunn JB, Nolte IM, Kamphuis P, Dinmohamed A, et al. Evolutionary landscape of clonal hematopoiesis in 3,359 individuals from the general population. Cancer Cell. 2023;41:1017–31.
Wilkinson ME, Charenton C, Nagai K. RNA splicing by the spliceosome. Annu Rev Biochem. 2020;89:359–88.
Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–9.
Zhang Y, Wu J, Qin T, Xu Z, Qu S, Pan L, et al. Comparison of the revised 4th (2016) and 5th (2022) editions of the World Health Organization classification of myelodysplastic neoplasms. Leukemia. 2022;36:2875–82.
Yip BH, Steeples V, Repapi E, Armstrong RN, Llorian M, Roy S, et al. The U2AF1S34F mutation induces lineage-specific splicing alterations in myelodysplastic syndromes. J Clin Invest. 2017;127:2206–21.
Kim SP, Srivatsan SN, Chavez M, Shirai CL, White BS, Ahmed T, et al. Mutant U2AF1-induced alternative splicing of H2afy (macroH2A1) regulates B-lymphopoiesis in mice. Cell Rep. 2021;36:109626.
Wheeler EC, Vora S, Mayer D, Kotini AG, Olszewska M, Park SS, et al. Integrative RNA-omics discovers GNAS alternative splicing as a phenotypic driver of splicing factor-mutant neoplasms. Cancer Discov. 2022;12:836–55.
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405.
Tolhurst G, Carter RN, Miller N, Mahaut-Smith MP. Purification of native bone marrow megakaryocytes for studies of gene expression. Methods Mol Biol. 2012;788:259–73.
Feng G, Gale RP, Cui W, Cai W, Huang G, Xu Z, et al. A systematic classification of megakaryocytic dysplasia and its impact on prognosis for patients with myelodysplastic syndromes. Exp Hematol Oncol. 2016;5:12.
Tomer A. Human marrow megakaryocyte differentiation: multiparameter correlative analysis identifies von Willebrand factor as a sensitive and distinctive marker for early (2N and 4N) megakaryocytes. Blood. 2004;104:2722–7.
Mazzi S, Lordier L, Debili N, Raslova H, Vainchenker W. Megakaryocyte and polyploidization. Exp Hematol. 2018;57:1–13.
Lordier L, Jalil A, Aurade F, Larbret F, Larghero J, Debili N, et al. Megakaryocyte endomitosis is a failure of late cytokinesis related to defects in the contractile ring and Rho/Rock signaling. Blood. 2008;112:3164–74.
Gao Y, Smith E, Ker E, Campbell P, Cheng E-c, Zou S, et al. Role of RhoA-specific guanine exchange factors in regulation of endomitosis in megakaryocytes. Dev Cell. 2012;22:573–84.
Yang X, Xu W, Hu Z, Zhang Y, Xu N. Chk1 is required for the metaphase–anaphase transition via regulating the expression and localization of Cdc20 and Mad2. Life Sci. 2014;106:12–8.
Mackay DR, Ullman KS, Solomon MJ. ATR and a Chk1-Aurora B pathway coordinate postmitotic genome surveillance with cytokinetic abscission. Mol Biol Cell. 2015;26:2217–26.
Michelena J, Gatti M, Teloni F, Imhof R, Altmeyer M. Basal CHK1 activity safeguards its stability to maintain intrinsic S-phase checkpoint functions. J Cell Biol. 2019;218:2865–75.
Neizer-Ashun F, Bhattacharya R. Reality CHEK: understanding the biology and clinical potential of CHK1. Cancer Lett. 2021;497:202–11.
Pellagatti A, Armstrong RN, Steeples V, Sharma E, Repapi E, Singh S, et al. Impact of spliceosome mutations on RNA splicing in myelodysplasia: dysregulated genes/pathways and clinical associations. Blood. 2018;132:1225–40.
Lee SC-W, North K, Kim E, Jang E, Obeng E, Lu SX, et al. Synthetic lethal and convergent biological effects of cancer-associated spliceosomal gene mutations. Cancer cell. 2018;34:225–41.
Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–95.
Moura PL, Mortera-Blanco T, Hofman IJ, Todisco G, Kretzschmar WW, Björklund A-C, et al. Erythroid differentiation enhances RNA mis-splicing in SF3B1-mutant myelodysplastic syndromes with ring sideroblasts. Cancer Res. 2024;84:211–25.
Hasserjian RP, Germing U, Malcovati L. Diagnosis and classification of myelodysplastic syndromes. Blood. 2023;142:2247–57.
Clough CA, Pangallo J, Sarchi M, Ilagan JO, North K, Bergantinos R, et al. Coordinated missplicing of TMEM14C and ABCB7 causes ring sideroblast formation in SF3B1-mutant myelodysplastic syndrome. Blood. 2022;139:2038–49.
Kim E, Ilagan Janine O, Liang Y, Daubner Gerrit M, Lee Stanley CW, Ramakrishnan A, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer cell. 2015;27:617–30.
Dalton WB, Helmenstine E, Walsh N, Gondek LP, Kelkar DS, Read A, et al. Hotspot SF3B1 mutations induce metabolic reprogramming and vulnerability to serine deprivation. J Clin Invest. 2019;129:4708–23.
Lappin KM, Barros EM, Jhujh SS, Irwin GW, McMillan H, Liberante FG, et al. Cancer-associated SF3B1 mutations confer a BRCA-like cellular phenotype and synthetic lethality to PARP inhibitors. Cancer Res. 2022;82:819–30.
Liu ZS, Sinha S, Bannister M, Song A, Arriaga-Gomez E, McKeeken AJ, et al. R-loop accumulation in spliceosome mutant leukemias confers sensitivity to PARP1 inhibition by triggering transcription-replication conflicts. Cancer Res. 2024;84:577–97.
Benbarche S, Pineda JMB, Galvis LB, Biswas J, Liu B, Wang E, et al. GPATCH8 modulates mutant SF3B1 mis-splicing and pathogenicity in hematologic malignancies. Mol Cell. 2024;84:1886–903.
Dolatshad H, Pellagatti A, Liberante FG, Llorian M, Repapi E, Steeples V, et al. Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia. 2016;30:2322–31.
Zhang J, Ali AM, Lieu YK, Liu Z, Gao J, Rabadan R, et al. Disease-causing mutations in SF3B1 alter splicing by disrupting interaction with SUGP1. Mol Cell. 2019;76:82–95.
Liu Z, Zhang J, Sun Y, Perea-Chamblee TE, Manley JL, Rabadan R. Pan-cancer analysis identifies mutations in SUGP1 that recapitulate mutant SF3B1 splicing dysregulation. Proc Natl Acad Sci USA. 2020;117:10305–12.
Mian SA, Philippe C, Maniati E, Protopapa P, Bergot T, Piganeau M, et al. Vitamin B5 and succinyl-CoA improve ineffective erythropoiesis in SF3B1-mutated myelodysplasia. Sci Transl Med. 2023;15:eabn5135.
Garcia-Ruiz C, Martínez-Valiente C, Cordón L, Liquori A, Fernández-González R, Pericuesta E, et al. Concurrent Zrsr2 mutation and Tet2 loss promote myelodysplastic neoplasm in mice. Leukemia. 2022;36:2509–18.
Park SM, Ou J, Chamberlain L, Simone TM, Yang H, Virbasius CM, et al. U2AF35(S34F) promotes transformation by directing aberrant ATG7 pre-mRNA 3’ end formation. Mol Cell. 2016;62:479–90.
Trakala M, Rodriguez-Acebes S, Maroto M, Symonds CE, Santamaria D, Ortega S, et al. Functional reprogramming of polyploidization in megakaryocytes. Dev Cell. 2015;32:155–67.
Sarchi M, Clough CA, Crosse EI, Kim J, Baquero Galvis LD, Aydinyan N, et al. Mis-splicing of mitotic regulators sensitizes SF3B1-mutated human HSCs to CHK1 inhibition. Blood Cancer Discov. 2024;5:353–70.
Machlus KR, Italiano JE Jr. The incredible journey: from megakaryocyte development to platelet formation. J Cell Biol. 2013;201:785–96.
Acknowledgements
We would like to express our sincere appreciation to Professors Omar Abdel-Wahab and Kevin Rouault-Pierre for their invaluable guidance and support throughout this research. Their expertise and constructive feedback have been instrumental in the development of this study.
Funding
This study is supported in part by National Natural Science Funds (Nos. 82170139, 81530008 and 82070134), CAMS Initiative Fund for Medical Sciences (Nos. 2023-I2M-2-007, 2022-I2M-1-022,), Clinical research fund from National Clinical Research Centre for Blood Diseases (Nos. 2023NCRCA0117 and 2023NCRCA0103), Medical and Healthy Science Innovative Program of Chinese Academy of Medical Science (2022-I2M-C&T-B-093).
Author information
Authors and Affiliations
Contributions
ZJX and BL initiated the concept for the study. ZJX and BL designed the study. WJZ performed the research, collected data, write and revised manuscript, JQL, BL, LY and YRY performed the research. TJQ, ZFX, QS, YJJ and HJW recruited patients and collected data. GH, LHS, HTW and JXZ provided support for the experiments. WJZ, BL and ZJX prepared the typescript. All authors read, approved the final manuscript, and agreed to submit for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Informed consent
The patient gave written informed consent compliant with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, W., Li, B., Liu, J. et al. Aberrant splicing of CHEK1 is a driver of megakaryocytic dysplasia in U2AF1S34F mutant myelodysplastic neoplasms. Leukemia 39, 2246–2255 (2025). https://doi.org/10.1038/s41375-025-02684-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41375-025-02684-6