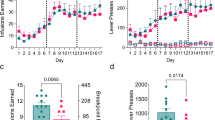
Abstract
Epigenetic mechanisms, like those involving DNA methylation, are thought to mediate the relationship between chronic cocaine dependence and molecular changes in addiction-related neurocircuitry, but have been understudied in human brain. We initially used reduced representation bisulfite sequencing (RRBS) to generate a methylome-wide profile of cocaine dependence in human post-mortem caudate tissue. We focused on the Iroquois Homeobox A (IRXA) gene cluster, where hypomethylation in exon 3 of IRX2 in neuronal nuclei was associated with cocaine dependence. We replicated this finding in an independent cohort and found similar results in the dorsal striatum from cocaine self-administering mice. Using epigenome editing and 3C assays, we demonstrated a causal relationship between methylation within the IRX2 gene body, CTCF protein binding, three-dimensional (3D) chromatin interaction, and gene expression. Together, these findings suggest that cocaine-related hypomethylation of IRX2 contributes to the development and maintenance of cocaine dependence through alterations in 3D chromatin structure in the caudate nucleus.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
American Psychiatric Association (APA). Diagnostic and Statistical Manual of Mental Disorders. 4th ed. Washington: American Psychiatric Association (APA); 2000.
Freeman WM, Lull ME, Patel KM, Brucklacher RM, Morgan D, Roberts DCS, et al. Gene expression changes in the medial prefrontal cortex and nucleus accumbens following abstinence from cocaine self-administration. BMC Neurosci. 2010;11:29.
Albertson D, Pruetz B, Schmidt C, Kuhn D, Kapatos G, Bannon M. Gene expression profile of the nucleus accumbens of human cocaine abusers: evidence for dysregulation of myelin. J Neurochem. 2004;88:1211–9.
Bannon MJ, Savonen CL, Jia H, Dachet F, Halter SD, Schmidt CJ, et al. Identification of long noncoding RNAs dysregulated in the midbrain of human cocaine abusers. J Neurochem. 2015;135:50–59.
Mash D, French-Mullen J, Adi N, Qin Y, Buck A, Pablo J, et al. Gene expression in human hippocampus from cocaine abusers identifies genes which regulate extracellular matrix remodeling. PLoS ONE. 2007;2:e1187.
Zhou Z, Yuan Q, Mash DC, Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc Natl Acad Sci USA. 2011;108:6626–31.
Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014;76 Part B:259–68.
Vaillancourt K, Ernst C, Mash D, Turecki G. DNA methylation dynamics and cocaine in the brain: progress and prospects. Genes. 2017;8:138.
Engmann O, Labonté B, Mitchell A, Bashtrykov P, Calipari ES, Rosenbluh C, et al. Cocaine-induced chromatin modifications associate with increased expression and three-dimensional looping of Auts2. Biol Psychiatry. 2017;82:794–805.
LaPlant Q, Vialou V, Covington HE, Dumitriu D, Feng J, Warren BL, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–43.
Garavan H, Pankiewicz J, Bloom A, Cho J-K, Sperry L, Ross TJ, et al. Cue-induced cocaine craving: neuroanatomical specificity for drug users and drug stimuli. Am J Psychiatry. 2000;157:1789–98.
Volkow ND, Wang G-J, Telang F, Fowler JS, Logan J, Childress A-R, et al. Cocaine cues and dopamine in dorsal striatum: mechanism of craving in cocaine addiction. J Neurosci. 2006;26:6583–8.
Belin D, Everitt B. Cocaine seeking habits depend upon dopamine-dependent serial connectivity linking the ventral with the dorsal striatum. Neuron 2008;57:432–41.
Everitt BJ, Robbins TW. From the ventral to the dorsal striatum: devolving views of their roles in drug addiction. Neurosci Biobehav Rev. 2013;37:1946–54.
Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–81.
Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–41.
Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–30.
Matsumoto K, Nishihara S, Kamimura M, Shiraishi T, Otoguro T, Uehara M, et al. The prepattern transcription factor Irx2, a target of the FGF8/MAP kinase cascade, is involved in cerebellum formation. Nat Neurosci. 2004;7:605–12.
Kozlenkov A, Roussos P, Timashpolsky A, Barbu M, Rudchenko S, Bibikova M, et al. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Res. 2014;42:109–27.
Rizzardi LF, Hickey PF, Rodriguez DiBlasi V, Tryggvadóttir R, Callahan CM, Idrizi A, et al. Neuronal brain-region-specific DNA methylation and chromatin accessibility are associated with neuropsychiatric trait heritability. Nat Neurosci. 2019;22:307–16.
Tena JJ, Alonso ME, de la Calle-Mustienes E, Splinter E, de Laat W, Manzanares M, et al. An evolutionarily conserved three-dimensional structure in the vertebrate Irx clusters facilitates enhancer sharing and coregulation. Nat Commun. 2011;2:310.
Hashimoto H, Wang D, Horton JR, Zhang X, Corces VG, Cheng X. Structural basis for the versatile and methylation-dependent binding of CTCF to DNA. Mol Cell. 2017;66:711–720.e3.
Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 2007;128:1231–45.
Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–812.
Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–28.
Bernácer J, Prensa L, Giménez-Amaya JM. Distribution of GABAergic interneurons and dopaminergic cells in the functional territories of the human striatum. PLoS ONE. 2012;7:e30504.
Tepper JM, Tecuapetla F, Koós T, Ibáñez-Sandoval O. Heterogeneity and diversity of striatal GABAergic interneurons. Front Neuroanat. 2010;4:150.
Lobo MK, Nestler EJ. The striatal balancing act in drug addiction: distinct roles of direct and indirect pathway medium spiny neurons. Front Neuroanat. 2011;5:41.
Yao P, Lin P, Gokoolparsadh A, Assareh A, Thang MWC, Voineagu I. Coexpression networks identify brain region–specific enhancer RNAs in the human brain. Nat Neurosci. 2015;18:1168–74.
Hannon E, Marzi SJ, Schalkwyk LS, Mill J. Genetic risk variants for brain disorders are enriched in cortical H3K27ac domains. Mol Brain. 2019;12:7.
Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–69.
Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010;466:253–7.
Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–90.
Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362:eaat8127.
Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15:R65.
Cates HM, Heller EA, Lardner CK, Purushothaman I, Peña CJ, Walker DM, et al. Transcription factor E2F3a in nucleus accumbens affects cocaine action via transcription and alternative splicing. Biol Psychiatry. 2018;84:167–79.
Baker-Andresen D, Zhao Q, Li X, Jupp B, Chesworth R, Lawrence AJ, et al. Persistent variations in neuronal DNA methylation following cocaine self-administration and protracted abstinence in mice. Neuroepigenetics. 2015;4:1–11.
Ahn J-I, Lee K-H, Shin D-M, Shim J-W, Lee J-S, Chang SY, et al. Comprehensive transcriptome analysis of differentiation of embryonic stem cells into midbrain and hindbrain neurons. Dev Biol. 2004;265:491–501.
Kasper C, Hebert FO, Aubin-Horth N, Taborsky B. Divergent brain gene expression profiles between alternative behavioural helper types in a cooperative breeder. Mol Ecol. 2018;27:4136–51.
Gomez-Velazquez M, Badia-Careaga C, Lechuga-Vieco AV, Nieto-Arellano R, Tena JJ, Rollan I, et al. CTCF counter-regulates cardiomyocyte development and maturation programs in the embryonic heart. PLoS Genet. 2017;13:e1006985.
Kelz MB, Chen J, Carlezon WA, Whisler K, Gilden L, Beckmann AM, et al. Expression of the transcription factor ΔFosB in the brain controls sensitivity to cocaine. Nature 1999;401:272–6.
Nestler EJ. The neurobiology of cocaine addiction. Sci Pract Perspect. 2005;3:4–10.
Bannon MJ, Johnson MM, Michelhaugh SK, Hartley ZJ, Halter SD, David JA, et al. A molecular profile of cocaine abuse includes the differential expression of genes that regulate transcription, chromatin, and dopamine cell phenotype. Neuropsychopharmacology 2014;39:1–9.
Cannella N, Oliveira AMM, Hemstedt T, Lissek T, Buechler E, Bading H, et al. Dnmt3a2 in the nucleus accumbens shell is required for reinstatement of cocaine seeking. J Neurosci. 2018;38:7516–28.
Chandra R, Francis TC, Konkalmatt P, Amgalan A, Gancarz AM, Dietz DM, et al. Opposing role for Egr3 in nucleus accumbens cell subtypes in cocaine action. J Neurosci. 2015;35:7927–37.
Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016;2016:baw100.
Saftig P, Lichtenthaler SF. The alpha secretase ADAM10: A metalloprotease with multiple functions in the brain. Prog Neurobiol. 2015;135:1–20.
Shukla M, Maitra S, Hernandez J-F, Govitrapong P, Vincent B. Methamphetamine regulates βAPP processing in human neuroblastoma cells. Neurosci Lett. 2019;701:20–5.
Maurano MT, Wang H, John S, Shafer A, Canfield T, Lee K, et al. Role of DNA methylation in modulating transcription factor occupancy. Cell Rep. 2015;12:1184–95.
Zhang TY, Hellstrom IC, Bagot RC, Wen X, Diorio J, Meaney MJ. Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. J Neurosci. 2010;30:13130–7.
Zhang TY, Keown CL, Wen X, Li J, Vousden DA, Anacker C, et al. Environmental enrichment increases transcriptional and epigenetic differentiation between mouse dorsal and ventral dentate gyrus. Nat Commun. 2018;9:1–11.
Gross JA, Fiori LM, Labonté B, Lopez JP, Turecki G. Effects of promoter methylation on increased expression of polyamine biosynthetic genes in suicide. J Psychiatr Res. 2013;47:513–9.
Iwata A, Nagata K, Hatsuta H, Takuma H, Bundo M, Iwamoto K, et al. Altered CpG methylation in sporadic Alzheimer’s disease is associated with APP and MAPT dysregulation. Hum Mol Genet. 2014;23:648–56.
Lutz PE, Tanti A, Gasecka A, Barnett-Burns S, Kim JJ, Zhou Y, et al. Association of a history of child abuse with impaired myelination in the anterior cingulate cortex: convergent epigenetic, transcriptional, and morphological evidence. Am J Psychiatry. 2017;174:1185–94.
Guo JU, Su Y, Shin JJH, Shin JJH, Li H, Xie B, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2013;17:215–22.
Li X, Zhao Q, Wei W, Lin Q, Magnan C, Emami MR, et al. The DNA modification N6-methyl-2’-deoxyadenosine (m6dA) drives activity-induced gene expression and is required for fear extinction. Nat Neurosci. 2019;22:534–44.
Feng J, Shao N, Szulwach KE, Vialou V, Huynh J, Zhong C, et al. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat Neurosci. 2015;18:536–44.
Luo C, Keown CL, Kurihara L, Zhou J, He Y, Li J, et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 2017;357:600–4.
Chen GG, Diallo AB, Poujol R, Nagy C, Staffa A, Vaillancourt K, et al. BisQC: an operational pipeline for multiplexed bisulfite sequencing. BMC Genomics. 2014;15:290.
Cavalcante RG, Sartor MA. Annotatr: genomic regions in context. Bioinformatics 2017;33:2381–3.
Sheffield NC, Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics 2016;32:587–9.
GTEx Consortium TGte. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–60.
Chen GG, Gross JA, Lutz P-E, Vaillancourt K, Maussion G, Bramoulle A, et al. Medium throughput bisulfite sequencing for accurate detection of 5-methylcytosine and 5-hydroxymethylcytosine. BMC Genomics. 2017;18:96.
Johnson AR, Thibeault KC, Lopez AJ, Peck EG, Sands LP, Sanders CM, et al. Cues play a critical role in estrous cycle-dependent enhancement of cocaine reinforcement. Neuropsychopharmacology. 2019; https://doi.org/10.1038/s41386-019-0320-0.
Ea V, Court F, Forne T. Quantitative analysis of intra-chromosomal contacts: the 3C-qPCR method. Methods Mol Biol. 2017;1589:75–88.
Braem C, Recolin B, Rancourt RC, Angiolini C, Barthès P, Branchu P, et al. Genomic matrix attachment region and chromosome conformation capture quantitative real time PCR assays identify novel putative regulatory elements at the imprinted Dlk1/Gtl2 locus. J Biol Chem. 2008;283:18612–20.
Acknowledgements
We are deeply grateful to the families of the subjects used in this study. We would also like to acknowledge the teams of technicians at the Miami Brain Endowment BankTM and the Douglas Bell Canada Brain Bank, and the bioinformaticians who have worked on these data (Alpha Diallo, Raphaël Poujol and Alexandre Bramoulle). This work was supported by a Canadian Institute of Health Research Doctoral Fellowship awarded to KV, National Institute of Drug Abuse Grants DA033684 awarded to GT and DCM, R00 DA04211 to ESC, and P01 DA047233 to EJN, and grants from the Whitehall Foundation, the Edward Mallinckrodt Jr., Foundation, and the Brain and Behavior Research Foundation to ESC.
Author information
Authors and Affiliations
Contributions
Manuscript preparation: KV; experimental design and data collection: KV, JY, GGC; data analysis: KV, CE, TF, J-FT, ZA; animal experiments: AL, KCT, BL; resources and support: EJN, ESC, CN, DCM, GT.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Vaillancourt, K., Yang, J., Chen, G.G. et al. Cocaine-related DNA methylation in caudate neurons alters 3D chromatin structure of the IRXA gene cluster. Mol Psychiatry 26, 3134–3151 (2021). https://doi.org/10.1038/s41380-020-00909-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41380-020-00909-x
This article is cited by
-
Incubation of methamphetamine craving in punishment-resistant individuals is associated with activation of specific gene networks in the rat dorsal striatum
Molecular Psychiatry (2024)
-
Multi-omics profiling of DNA methylation and gene expression alterations in human cocaine use disorder
Translational Psychiatry (2024)
-
Cis-meQTL for cocaine use-associated DNA methylation in an HIV-positive cohort show pleiotropic effects on multiple traits
BMC Genomics (2023)
-
Mechanisms underlying microRNA-222-3p modulation of methamphetamine-induced conditioned place preference in the nucleus accumbens in mice
Psychopharmacology (2022)
-
Hippocampal Cannabinoid 1 Receptors Are Modulated Following Cocaine Self-administration in Male Rats
Molecular Neurobiology (2022)