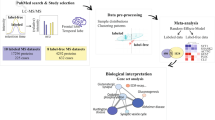
Abstract
Pathophysiological evolutions in early-stage Alzheimer’s disease (AD) are not well understood. We used data of 2923 Olink plasma proteins from 51,296 non-demented middle-aged adults. During a follow-up of 15 years, 689 incident AD cases occurred. Cox-proportional hazard models were applied to identify AD-associated proteins in different time intervals. Through linking to protein categories, changing sequences of protein z-scores can reflect pathophysiological evolutions. Mendelian randomization using blood protein quantitative loci data provided causal evidence for potentially druggable proteins. We identified 48 AD-related proteins, with CEND1, GFAP, NEFL, and SYT1 being top hits in both near-term (HR:1.15–1.77; P:9.11 × 10−65–2.78 × 10−6) and long-term AD risk (HR:1.20-1.54; P:2.43 × 10−21–3.95 × 10−6). These four proteins increased 15 years before AD diagnosis and progressively escalated, indicating early and sustained dysfunction in synapse and neurons. Proteins related to extracellular matrix organization, apoptosis, innate immunity, coagulation, and lipid homeostasis showed early disturbances, followed by malfunctions in metabolism, adaptive immunity, and final synaptic and neuronal loss. Combining CEND1, GFAP, NEFL, and SYT1 with demographics generated desirable predictions for 10-year (AUC = 0.901) and over-10-year AD (AUC = 0.864), comparable to full model. Mendelian randomization supports potential genetic link between CEND1, SYT1, and AD as outcome. Our findings highlight the importance of exploring the pathophysiological evolutions in early stages of AD, which is essential for the development of early biomarkers and precision therapeutics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The main data, including the individual-level phenotypic and genetic data used in this study were accessed from the UK Biobank under the application number 19542 and are available through UKB (https://www.ukbiobank.ac.uk/).
Code availability
Analyses were performed using R software (v4.3.1). We considered the two-tailed P < 0.05 significant. Code is available from the corresponding author by request.
References
Scheltens P, De Strooper B, Kivipelto M, Holstege H, Chetelat G, Teunissen CE, et al. Alzheimer’s disease. Lancet. 2021;397:1577–90.
Bai B, Vanderwall D, Li Y, Wang X, Poudel S, Wang H, et al. Proteomic landscape of Alzheimer’s Disease: novel insights into pathogenesis and biomarker discovery. Mol Neurodegener. 2021;16:55.
Jack CR Jr, Holtzman DM. Biomarker modeling of Alzheimer’s disease. Neuron. 2013;80:1347–58.
Hampel H, Cummings J, Blennow K, Gao P, Jack CR Jr, Vergallo A. Developing the ATX(N) classification for use across the Alzheimer disease continuum. Nat Rev Neurol. 2021;17:580–9.
Sun BB, Chiou J, Traylor M, Benner C, Hsu YH, Richardson TG, et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature. 2023;622:329–38.
Walker KA, Chen J, Zhang J, Fornage M, Yang Y, Zhou L, et al. Large-scale plasma proteomic analysis identifies proteins and pathways associated with dementia risk. Nat Aging. 2021;1:473–89.
Higginbotham L, Ping L, Dammer EB, Duong DM, Zhou M, Gearing M, et al. Integrated proteomics reveals brain-based cerebrospinal fluid biomarkers in asymptomatic and symptomatic Alzheimer’s disease. Science Advances. 2020;6:eaaz9360%L 9361.
Johnson ECB, Bian S, Haque RU, Carter EK, Watson CM, Gordon BA, et al. Cerebrospinal fluid proteomics define the natural history of autosomal dominant Alzheimer’s disease. Nat Med. 2023;29:1979–88.
Bai B, Wang X, Li Y, Chen PC, Yu K, Dey KK, et al. Deep Multilayer Brain Proteomics Identifies Molecular Networks in Alzheimer’s Disease Progression. Neuron. 2020;105:975–991 e977.
Johnson ECB, Dammer EB, Duong DM, Ping L, Zhou M, Yin L, et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med. 2020;26:769–80.
Johnson ECB, Carter EK, Dammer EB, Duong DM, Gerasimov ES, Liu Y, et al. Large-scale deep multi-layer analysis of Alzheimer’s disease brain reveals strong proteomic disease-related changes not observed at the RNA level. Nat Neurosci. 2022;25:213–25.
Jiang Y, Zhou X, Ip FC, Chan P, Chen Y, Lai NCH, et al. Large-scale plasma proteomic profiling identifies a high-performance biomarker panel for Alzheimer’s disease screening and staging. Alzheimers Dement. 2022;18:88–102.
Walker KA, Chen J, Shi L, Yang Y, Fornage M, Zhou L, et al. Proteomics analysis of plasma from middle-aged adults identifies protein markers of dementia risk in later life. Sci Transl Med. 2023;15:eadf5681.
Tanaka T, Lavery R, Varma V, Fantoni G, Colpo M, Thambisetty M. et al. Plasma proteomic signatures predict dementia and cognitive impairment. Alzheimers Dement (N Y.). 2020;6:e12018.
Hye A, Lynham S, Thambisetty M, Causevic M, Campbell J, Byers HL, et al. Proteome-based plasma biomarkers for Alzheimer’s disease. Brain. 2006;129:3042–50.
de Sousa Maciel I, Piironen A-K, Afonin AM, Ivanova M, Alatalo A, Jadhav KK, et al. Plasma proteomics discovery of mental health risk biomarkers in adolescents. Nature Mental Health. 2023;1:596–605.
Consortium TG, Aguet F, Anand S, Ardlie KG, Gabriel S, Getz GA, et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–30.
Milà-Alomà M, Ashton NJ, Shekari M, Salvadó G, Ortiz-Romero P, Montoliu-Gaya L, et al. Plasma p-tau231 and p-tau217 as state markers of amyloid-β pathology in preclinical Alzheimer’s disease. Nat Med. 2022;28:1797–801.
Mila-Aloma M, Salvado G, Gispert JD, Vilor-Tejedor N, Grau-Rivera O, Sala-Vila A, et al. Amyloid beta, tau, synaptic, neurodegeneration, and glial biomarkers in the preclinical stage of the Alzheimer’s continuum. Alzheimers Dement. 2020;16:1358–71.
Guo Y, Shen XN, Wang HF, Chen SD, Zhang YR, Chen SF, et al. The dynamics of plasma biomarkers across the Alzheimer’s continuum. Alzheimers Res Ther. 2023;15:31.
Johansson C, Thordardottir S, Laffita-Mesa J, Rodriguez-Vieitez E, Zetterberg H, Blennow K, et al. Plasma biomarker profiles in autosomal dominant Alzheimer’s disease. Brain. 2023;146:1132–40.
Del Campo M, Peeters CFW, Johnson ECB, Vermunt L, Hok AHYS, van Nee M, et al. CSF proteome profiling across the Alzheimer’s disease spectrum reflects the multifactorial nature of the disease and identifies specific biomarker panels. Nat Aging. 2022;2:1040–53.
Ikonomovic MD, Abrahamson EE, Isanski BA, Wuu J, Mufson EJ, DeKosky ST. Superior frontal cortex cholinergic axon density in mild cognitive impairment and early Alzheimer disease. Arch Neurol. 2007;64:1312–7.
Chang YT, Huang CW, Chen NC, Lin KJ, Huang SH, Chang WN, et al. Hippocampal Amyloid Burden with Downstream Fusiform Gyrus Atrophy Correlate with Face Matching Task Scores in Early Stage Alzheimer’s Disease. Front Aging Neurosci. 2016;8:145.
Koch G, Casula EP, Bonnì S, Borghi I, Assogna M, Minei M, et al. Precuneus magnetic stimulation for Alzheimer’s disease: a randomized, sham-controlled trial. Brain. 2022;145:3776–86.
Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51:414–30.
Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner K, et al. FinnGen: Unique genetic insights from combining isolated population and national health register data. medRxiv 2022: 2022.2003.2003.22271360.
Benedet AL, Milà-Alomà M, Vrillon A, Ashton NJ, Pascoal TA, Lussier F, et al. Differences Between Plasma and Cerebrospinal Fluid Glial Fibrillary Acidic Protein Levels Across the Alzheimer Disease Continuum. JAMA Neurol. 2021;78:1471–83.
Cicognola C, Janelidze S, Hertze J, Zetterberg H, Blennow K, Mattsson-Carlgren N, et al. Plasma glial fibrillary acidic protein detects Alzheimer pathology and predicts future conversion to Alzheimer dementia in patients with mild cognitive impairment. Alzheimers Res Ther. 2021;13:68.
de Wolf F, Ghanbari M, Licher S, McRae-McKee K, Gras L, Weverling GJ, et al. Plasma tau, neurofilament light chain and amyloid-beta levels and risk of dementia; a population-based cohort study. Brain. 2020;143:1220–32.
Guo Y, You J, Zhang Y, Liu WS, Huang YY, Zhang YR, et al. Plasma proteomic profiles predict future dementia in healthy adults. Nat Aging. 2024;4:247–60.
Quinn JP, Kandigian SE, Trombetta BA, Arnold SE, Carlyle BC. VGF as a biomarker and therapeutic target in neurodegenerative and psychiatric diseases. Brain Commun. 2021;3:fcab261.
Wingo AP, Dammer EB, Breen MS, Logsdon BA, Duong DM, Troncosco JC, et al. Large-scale proteomic analysis of human brain identifies proteins associated with cognitive trajectory in advanced age. Nat Commun. 2019;10:1619.
Eldjarn GH, Ferkingstad E, Lund SH, Helgason H, Magnusson OT, Gunnarsdottir K, et al. Large-scale plasma proteomics comparisons through genetics and disease associations. Nature. 2023;622:348–58.
Dammer EB, Ping L, Duong DM, Modeste ES, Seyfried NT, Lah JJ, et al. Multi-platform proteomic analysis of Alzheimer’s disease cerebrospinal fluid and plasma reveals network biomarkers associated with proteostasis and the matrisome. Alzheimers Res Ther. 2022;14:174.
Livingston G, Huntley J, Liu KY, Costafreda SG, Selbaek G, Alladi S, et al. Dementia prevention, intervention, and care: 2024 report of the Lancet standing Commission. Lancet. 2024;404:572–628.
Rosenzweig N, Kleemann KL, Rust T, Carpenter M, Grucci M, Aronchik M, et al. Sex-dependent APOE4 neutrophil-microglia interactions drive cognitive impairment in Alzheimer’s disease. Nat Med. 2024;30:2990–3003.
Brinkmalm A, Larsson V, Janelidze S, Zetterberg H, Blennow K, Hansson O. Cerebrospinal fluid levels of SNAP-25 and SYT1 in Alzheimer’s and Parkinson’s disease. Alzheimer’s & Dementia. 2020;16:e044515.
Xie W, Guo D, Li J, Yue L, Kang Q, Chen G, et al. CEND1 deficiency induces mitochondrial dysfunction and cognitive impairment in Alzheimer’s disease. Cell Death Differ. 2022;29:2417–28.
Chen X, Holtzman DM. Emerging roles of innate and adaptive immunity in Alzheimer’s disease. Immunity. 2022;55:2236–54.
Obulesu M, Lakshmi MJ. Apoptosis in Alzheimer’s disease: an understanding of the physiology, pathology and therapeutic avenues. Neurochem Res. 2014;39:2301–12.
Gunturkun O, Strockens F, Ocklenburg S. Brain Lateralization: A Comparative Perspective. Physiol Rev. 2020;100:1019–63.
Ocklenburg S, Guo ZV. Cross-hemispheric communication: Insights on lateralized brain functions. Neuron. 2024;112:1222–34.
Mesulam MM. Temporopolar regions of the human brain. Brain. 2023;146:20–41.
Banks SJ, Zhuang X, Bayram E, Bird C, Cordes D, Caldwell JZK, et al. Default Mode Network Lateralization and Memory in Healthy Aging and Alzheimer’s Disease. J Alzheimers Dis. 2018;66:1223–34.
Jack CR Jr, Andrews JS, Beach TG, Buracchio T, Dunn B, Graf A, et al. Revised criteria for diagnosis and staging of Alzheimer’s disease: Alzheimer’s Association Workgroup. Alzheimers Dement. 2024;20:5143–69.
Fjell AM, McEvoy L, Holland D, Dale AM, Walhovd KB. Alzheimer’s Disease Neuroimaging I. What is normal in normal aging? Effects of aging, amyloid and Alzheimer’s disease on the cerebral cortex and the hippocampus. Prog Neurobiol. 2014;117:20–40.
Öhrfelt A, Brinkmalm A, Dumurgier J, Brinkmalm G, Hansson O, Zetterberg H, et al. The pre-synaptic vesicle protein synaptotagmin is a novel biomarker for Alzheimer’s disease. Alzheimers Res Ther. 2016;8:41.
T P, Katta B, Lulu SS, Sundararajan V Gene expression analysis reveals GRIN1, SYT1, and SYN2 as significant therapeutic targets and drug repurposing reveals lorazepam and lorediplon as potent inhibitors to manage Alzheimer’s disease. J Biomol Struct Dyn. 2023;1–22.
Wang H, Dey KK, Chen PC, Li Y, Niu M, Cho JH, et al. Integrated analysis of ultra-deep proteomes in cortex, cerebrospinal fluid and serum reveals a mitochondrial signature in Alzheimer’s disease. Mol Neurodegener. 2020;15:43.
Simonet C, Bestwick J, Jitlal M, Waters S, Ben-Joseph A, Marshall CR, et al. Assessment of Risk Factors and Early Presentations of Parkinson Disease in Primary Care in a Diverse UK Population. JAMA Neurol. 2022;79:359–69.
Williams SA, Kivimaki M, Langenberg C, Hingorani AD, Casas JP, Bouchard C, et al. Plasma protein patterns as comprehensive indicators of health. Nat Med. 2019;25:1851–7.
Oh HS, Rutledge J, Nachun D, Pálovics R, Abiose O, Moran-Losada P, et al. Organ aging signatures in the plasma proteome track health and disease. Nature. 2023;624:164–72.
Wik L, Nordberg N, Broberg J, Björkesten J, Assarsson E, Henriksson S, et al. Proximity Extension Assay in Combination with Next-Generation Sequencing for High-throughput Proteome-wide Analysis. Mol Cell Proteomics. 2021;20:100168.
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141.
Yu G. Gene Ontology Semantic Similarity Analysis Using GOSemSim. Methods Mol Biol. 2020;2117:207–15.
Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010;26:976–8.
Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–9.
Lindbohm JV, Mars N, Walker KA, Singh-Manoux A, Livingston G, Brunner EJ, et al. Plasma proteins, cognitive decline, and 20-year risk of dementia in the Whitehall II and Atherosclerosis Risk in Communities studies. Alzheimers Dement. 2022;18:612–24.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Lehallier B, Gate D, Schaum N, Nanasi T, Lee SE, Yousef H, et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat Med. 2019;25:1843–50.
Piehl N, van Olst L, Ramakrishnan A, Teregulova V, Simonton B, Zhang Z, et al. Cerebrospinal fluid immune dysregulation during healthy brain aging and cognitive impairment. Cell. 2022;185:5028–5039.e5013.
Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–80.
Miller KL, Alfaro-Almagro F, Bangerter NK, Thomas DL, Yacoub E, Xu J, et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci. 2016;19:1523–36.
Alfaro-Almagro F, Jenkinson M, Bangerter NK, Andersson JLR, Griffanti L, Douaud G, et al. Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage. 2018;166:400–24.
Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–55.
Ganna A, Ingelsson E. 5 year mortality predictors in 498,103 UK Biobank participants: a prospective population-based study. Lancet. 2015;386:533–40.
DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45.
Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018; 7.
Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383.
Liu B, Gloudemans MJ, Rao AS, Ingelsson E, Montgomery SB. Abundant associations with gene expression complicate GWAS follow-up. Nat Genet. 2019;51:768–9.
Sakaue S, Okada Y. GREP: genome for REPositioning drugs. Bioinformatics. 2019;35:3821–3.
Funding
National Key R&D Program of China 2023YFC3605400. Science and Technology Innovation 2030 Major Projects 2022ZD0211600. National Natural Science Foundation of China 82071201, 82271471, 82472055, 82071997, 92249305, 82402381, and 8247070942. Shanghai Municipal Science and Technology Major Project 2018SHZDZX01 and 2023SHZDZX02. Research Start-up Fund of Huashan Hospital 2022QD002. Excellence 2025 Talent Cultivation Program at Fudan University 3030277001. Program of Shanghai Academic Research Leader 23XD1420400. Shanghai Rising-Star Program 21QA1408700. Shanghai Municipal Science and Technology Major Project 2023SHZDZX02. Shanghai Municipal Health Commission Emerging Interdisciplinary Research Project 2022JC014. National Postdoctoral Program for Innovative Talents (BX20230087 to S.-D.C., BX20230089 to Y.-R.Z., and BX20240073 to Y.G.), Shanghai Pujiang Talent Program (23PJD006 to J.Y.). We want to thank all the participants and researchers from the UK Biobank.
Author information
Authors and Affiliations
Contributions
Conceptualization: JTY, WC. Methodology: JTY, WC, YZ, LBW, JY, YG, YH, XYH, YL. Investigation: JTY, WC, JFF, QD. Visualization: YZ, YG, YH, JY. Supervision: JTY, WC, JFF, QD. Writing—original draft: YZ, YG, YH, JY. Writing—review & editing: All authors
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The study was conducted following the Declaration of Helsinki. The UK Biobank has research tissue bank approval from the North West Multi-Center Research Ethics Committee (11/NW/0382). Written informed consent was obtained from all participants.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Guo, Y., He, Y. et al. Large-scale proteomic analyses of incident Alzheimer’s disease reveal new pathophysiological insights and potential therapeutic targets. Mol Psychiatry 30, 2347–2361 (2025). https://doi.org/10.1038/s41380-024-02840-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41380-024-02840-x
This article is cited by
-
Biomarker identification for Alzheimer’s disease through integration of comprehensive Mendelian randomization and proteomics data
Journal of Translational Medicine (2025)
-
Alzheimer’s disease patient brain extracts induce multiple pathologies in novel vascularized neuroimmune organoids for disease modeling and drug discovery
Molecular Psychiatry (2025)