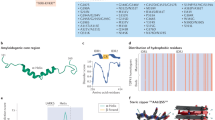
Abstract
Alzheimer’s disease (AD) and Alzheimer’s disease-related dementias (ADRDs) are among the most prevalent neurodegenerative diseases, characterized by progressive cognitive decline driven by complex and overlapping pathological mechanisms. While amyloid plaques, neurofibrillary tangles, and Lewy bodies are well-established hallmarks, TAR DNA-binding protein 43 (TDP-43) pathology has emerged as a critical contributor to disease progression, particularly in cases exhibiting hippocampal sclerosis and severe brain atrophy. TDP-43 pathology is defined by its cytoplasmic mislocalization, aberrant aggregation, and nuclear depletion, leading to disruptions in RNA metabolism, stress granule dynamics, and mitochondrial function. Increasing evidence suggests that TDP-43 pathology not only exacerbates neuronal degeneration but also interacts with Aβ plaques, tau tangles, and α-synuclein aggregates, compounding neurodegenerative processes and accelerating cognitive decline. Despite its growing recognition, TDP-43 pathology remains underexplored compared to other proteinopathies in AD and ADRDs, highlighting the need for further mechanistic studies and targeted therapeutic development. In this review, we summarize the current understanding of TDP-43 pathology in AD and ADRDs, with a focus on its role in disease progression. We further discuss the molecular mechanisms underlying TDP-43-associated neurodegeneration in AD and ADRDs, emphasizing RNA dysregulation, mitochondrial dysfunction, disrupted protein homeostasis, stress response alternations, and nuclear-cytoplasmic transport impairments. Lastly, given the significant impact on disease pathology, we review ongoing efforts to treat TDP-43-associated neurodegeneration, including antisense oligonucleotides, small-molecule inhibitors, and peptide-based interventions aimed at restoring TDP-43 function or preventing its neurotoxicity and pathological aggregation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Baumgart M, Snyder HM, Carrillo MC, Fazio S, Kim H, Johns H. Summary of the evidence on modifiable risk factors for cognitive decline and dementia: a population-based perspective. Alzheimers Dement. 2015;11:718–26.
Livingston G, Huntley J, Liu KY, Costafreda SG, Selbaek G, Alladi S, et al. Dementia prevention, intervention, and care: 2024 report of the lancet standing commission. Lancet. 2024;404:572–628.
2024 Alzheimer’s disease facts and figures. Alzheimers Dement. 2024;20:3708–821.
Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–96.
Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005;280:37572–84.
Wang HY, Wang IF, Bose J, Shen CK. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–9.
Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3.
Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–11.
Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–4.
Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–72.
Cairns NJ, Neumann M, Bigio EH, Holm IE, Troost D, Hatanpaa KJ, et al. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am J Pathol. 2007;171:227–40.
Amador-Ortiz C, Lin WL, Ahmed Z, Personett D, Davies P, Duara R, et al. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann Neurol. 2007;61:435–45.
Chanson JB, Echaniz-Laguna A, Vogel T, Mohr M, Benoilid A, Kaltenbach G, et al. TDP43-positive intraneuronal inclusions in a patient with motor neuron disease and Parkinson’s disease. Neurodegener Dis. 2010;7:260–4.
Schwab C, Arai T, Hasegawa M, Yu S, McGeer PL. Colocalization of transactivation-responsive DNA-binding protein 43 and huntingtin in inclusions of Huntington disease. J Neuropathol Exp Neurol. 2008;67:1159–65.
Higashi S, Iseki E, Yamamoto R, Minegishi M, Hino H, Fujisawa K, et al. Concurrence of TDP-43, tau and alpha-synuclein pathology in brains of Alzheimer’s disease and dementia with Lewy bodies. Brain Res. 2007;1184:284–94.
Weihl CC, Temiz P, Miller SE, Watts G, Smith C, Forman M, et al. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J Neurol Neurosurg Psychiatry. 2008;79:1186–9.
Nelson PT, Trojanowski JQ, Abner EL, Al-Janabi OM, Jicha GA, Schmitt FA, et al. “New Old Pathologies”: AD, PART, and cerebral age-related TDP-43 with sclerosis (CARTS). J Neuropathol Exp Neurol. 2016;75:482–98.
Nelson PT, Dickson DW, Trojanowski JQ, Jack CR, Boyle PA, Arfanakis K, et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain. 2019;142:1503–27.
Raji CA, Lopez OL, Kuller LH, Carmichael OT, Becker JT. Age, Alzheimer disease, and brain structure. Neurology. 2009;73:1899–905.
Smith MA. Alzheimer disease. Int Rev Neurobiol. 1998;42:1–54.
Zhu X, Lee HG, Casadesus G, Avila J, Drew K, Perry G, et al. Oxidative imbalance in Alzheimer’s disease. Mol Neurobiol. 2005;31:205–17.
Czech C, Tremp G, Pradier L. Presenilins and Alzheimer’s disease: biological functions and pathogenic mechanisms. Prog Neurobiol. 2000;60:363–84.
Tanahashi H, Tabira T. Alzheimer’s disease-associated presenilin 2 interacts with DRAL, an LIM-domain protein. Hum Mol Genet. 2000;9:2281–9.
Bellenguez C, Kucukali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Genet. 2022;54:412–36.
DeTure MA, Dickson DW. The neuropathological diagnosis of Alzheimer’s disease. Mol Neurodegener. 2019;14:32.
Josephs KA, Murray ME, Whitwell JL, Parisi JE, Petrucelli L, Jack CR, et al. Staging TDP-43 pathology in Alzheimer’s disease. Acta Neuropathol. 2014;127:441–50.
Josephs KA, Murray ME, Whitwell JL, Tosakulwong N, Weigand SD, Petrucelli L, et al. Updated TDP-43 in Alzheimer’s disease staging scheme. Acta Neuropathol. 2016;131:571–85.
James BD, Wilson RS, Boyle PA, Trojanowski JQ, Bennett DA, Schneider JA. TDP-43 stage, mixed pathologies, and clinical Alzheimer’s-type dementia. Brain. 2016;139:2983–93.
Gomes LA, Hipp SA, Rijal Upadhaya A, Balakrishnan K, Ospitalieri S, Koper MJ, et al. Abeta-induced acceleration of Alzheimer-related tau-pathology spreading and its association with prion protein. Acta Neuropathol. 2019;138:913–41.
Robinson JL, Lee EB, Xie SX, Rennert L, Suh E, Bredenberg C, et al. Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain. 2018;141:2181–93.
Tome SO, Vandenberghe R, Ospitalieri S, Van Schoor E, Tousseyn T, Otto M, et al. Distinct molecular patterns of TDP-43 pathology in Alzheimer’s disease: relationship with clinical phenotypes. Acta Neuropathol Commun. 2020;8:61.
Katsumata Y, Fardo DW, Kukull WA, Nelson PT. Dichotomous scoring of TDP-43 proteinopathy from specific brain regions in 27 academic research centers: associations with Alzheimer’s disease and cerebrovascular disease pathologies. Acta Neuropathol Commun. 2018;6:142.
Power MC, Mormino E, Soldan A, James BD, Yu L, Armstrong NM, et al. Combined neuropathological pathways account for age-related risk of dementia. Ann Neurol. 2018;84:10–22.
Uryu K, Nakashima-Yasuda H, Forman MS, Kwong LK, Clark CM, Grossman M, et al. Concomitant TAR-DNA-binding protein 43 pathology is present in Alzheimer disease and corticobasal degeneration but not in other tauopathies. J Neuropathol Exp Neurol. 2008;67:555–64.
Nelson PT, Lee EB, Cykowski MD, Alafuzoff I, Arfanakis K, Attems J, et al. LATE-NC staging in routine neuropathologic diagnosis: an update. Acta Neuropathol. 2023;145:159–73.
Katsumata Y, Wu X, Aung KZ, Fardo DW, Woodworth DC, Sajjadi SA, et al. Pure LATE-NC: Frequency, clinical impact, and the importance of considering APOE genotype when assessing this and other subtypes of non-Alzheimer’s pathologies. Acta Neuropathol. 2024;148:66.
Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–54.
McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ, et al. Clinical and pathological diagnosis of frontotemporal dementia: report of the work group on frontotemporal dementia and Pick’s disease. Arch Neurol. 2001;58:1803–9.
Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol. 2009;117:15–18.
Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, et al. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 2010;119:1–4.
Irwin DJ, Cairns NJ, Grossman M, McMillan CT, Lee EB, Van Deerlin VM, et al. Frontotemporal lobar degeneration: defining phenotypic diversity through personalized medicine. Acta Neuropathol. 2015;129:469–91.
Rademakers R, Neumann M, Mackenzie IR. Advances in understanding the molecular basis of frontotemporal dementia. Nat Rev Neurol. 2012;8:423–34.
Sampathu DM, Neumann M, Kwong LK, Chou TT, Micsenyi M, Truax A, et al. Pathological heterogeneity of frontotemporal lobar degeneration with ubiquitin-positive inclusions delineated by ubiquitin immunohistochemistry and novel monoclonal antibodies. Am J Pathol. 2006;169:1343–52.
Mackenzie IR, Baborie A, Pickering-Brown S, Du Plessis D, Jaros E, Perry RH, et al. Heterogeneity of ubiquitin pathology in frontotemporal lobar degeneration: classification and relation to clinical phenotype. Acta Neuropathol. 2006;112:539–49.
Mackenzie IR, Neumann M, Baborie A, Sampathu DM, Du Plessis D, Jaros E, et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011;122:111–3.
Lee EB, Porta S, Michael Baer G, Xu Y, Suh E, Kwong LK, et al. Expansion of the classification of FTLD-TDP: distinct pathology associated with rapidly progressive frontotemporal degeneration. Acta Neuropathol. 2017;134:65–78.
Tan RH, Guennewig B, Dobson-Stone C, Kwok JBJ, Kril JJ, Kiernan MC, et al. The underacknowledged PPA-ALS: a unique clinicopathologic subtype with strong heritability. Neurology. 2019;92:e1354–e1366.
Clark CN, Quaegebeur A, Nirmalananthan N, MacKinnon AD, Revesz T, Holton JL, et al. Foix-Chavany-Marie syndrome due to type E TDP43 pathology. Neuropathol Appl Neurobiol. 2020;46:292–5.
Koga S, Zhou X, Murakami A, Fernandez De Castro C, Baker MC, Rademakers R, et al. Concurrent tau pathologies in frontotemporal lobar degeneration with TDP-43 pathology. Neuropathol Appl Neurobiol. 2022;48:e12778.
Cykowski MD, Powell SZ, Schulz PE, Takei H, Rivera AL, Jackson RE, et al. Hippocampal sclerosis in older patients: practical examples and guidance with a focus on cerebral age-related TDP-43 with sclerosis. Arch Pathol Lab Med. 2017;141:1113–26.
Amador-Ortiz C, Ahmed Z, Zehr C, Dickson DW. Hippocampal sclerosis dementia differs from hippocampal sclerosis in frontal lobe degeneration. Acta Neuropathol. 2007;113:245–52.
Dickson DW, Baker M, Rademakers R. Common variant in GRN is a genetic risk factor for hippocampal sclerosis in the elderly. Neurodegener Dis. 2010;7:170–4.
Murray ME, Cannon A, Graff-Radford NR, Liesinger AM, Rutherford NJ, Ross OA, et al. Differential clinicopathologic and genetic features of late-onset amnestic dementias. Acta Neuropathol. 2014;128:411–21.
Nelson PT, Estus S, Abner EL, Parikh I, Malik M, Neltner JH, et al. ABCC9 gene polymorphism is associated with hippocampal sclerosis of aging pathology. Acta Neuropathol. 2014;127:825–43.
Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014;10:e1004606.
Nelson PT, Schmitt FA, Lin Y, Abner EL, Jicha GA, Patel E, et al. Hippocampal sclerosis in advanced age: clinical and pathological features. Brain. 2011;134:1506–18.
Lee EB, Lee VM, Trojanowski JQ, Neumann M. TDP-43 immunoreactivity in anoxic, ischemic and neoplastic lesions of the central nervous system. Acta Neuropathol. 2008;115:305–11.
McKee AC, Cairns NJ, Dickson DW, Folkerth RD, Keene CD, Litvan I, et al. The first NINDS/NIBIB consensus meeting to define neuropathological criteria for the diagnosis of chronic traumatic encephalopathy. Acta Neuropathol. 2016;131:75–86.
Johnson VE, Stewart W, Trojanowski JQ, Smith DH. Acute and chronically increased immunoreactivity to phosphorylation-independent but not pathological TDP-43 after a single traumatic brain injury in humans. Acta Neuropathol. 2011;122:715–26.
Burton EJ, Karas G, Paling SM, Barber R, Williams ED, Ballard CG, et al. Patterns of cerebral atrophy in dementia with Lewy bodies using voxel-based morphometry. Neuroimage. 2002;17:618–30.
Burton EJ, Barber R, Mukaetova-Ladinska EB, Robson J, Perry RH, Jaros E, et al. Medial temporal lobe atrophy on MRI differentiates Alzheimer’s disease from dementia with Lewy bodies and vascular cognitive impairment: a prospective study with pathological verification of diagnosis. Brain. 2009;132:195–203.
Outeiro TF, Koss DJ, Erskine D, Walker L, Kurzawa-Akanbi M, Burn D, et al. Dementia with Lewy bodies: an update and outlook. Mol Neurodegener. 2019;14:5.
McKeith IG, Boeve BF, Dickson DW, Halliday G, Taylor JP, Weintraub D, et al. Diagnosis and management of dementia with Lewy bodies: fourth consensus report of the DLB consortium. Neurology. 2017;89:88–100.
Serpell LC, Berriman J, Jakes R, Goedert M, Crowther RA. Fiber diffraction of synthetic alpha-synuclein filaments shows amyloid-like cross-beta conformation. Proc Natl Acad Sci USA. 2000;97:4897–902.
Goedert M, Spillantini MG, Del Tredici K, Braak H. 100 years of Lewy pathology. Nat Rev Neurol. 2013;9:13–24.
Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc Natl Acad Sci USA. 1998;95:6469–73.
Keogh MJ, Kurzawa-Akanbi M, Griffin H, Douroudis K, Ayers KL, Hussein RI, et al. Exome sequencing in dementia with Lewy bodies. Transl Psychiatry. 2016;6:e728.
Bras J, Guerreiro R, Darwent L, Parkkinen L, Ansorge O, Escott-Price V, et al. Genetic analysis implicates APOE, SNCA and suggests lysosomal dysfunction in the etiology of dementia with Lewy bodies. Hum Mol Genet. 2014;23:6139–46.
Guerreiro R, Ross OA, Kun-Rodrigues C, Hernandez DG, Orme T, Eicher JD, et al. Investigating the genetic architecture of dementia with Lewy bodies: a two-stage genome-wide association study. Lancet Neurol. 2018;17:64–74.
Vergouw LJM, van Steenoven I, van de Berg WDJ, Teunissen CE, van Swieten JC, Bonifati V, et al. An update on the genetics of dementia with Lewy bodies. Parkinsonism Relat Disord. 2017;43:1–8.
Nalls MA, Duran R, Lopez G, Kurzawa-Akanbi M, McKeith IG, Chinnery PF, et al. A multicenter study of glucocerebrosidase mutations in dementia with Lewy bodies. JAMA Neurol. 2013;70:727–35.
Heckman MG, Soto-Ortolaza AI, Contreras MYS, Murray ME, Pedraza O, Diehl NN, et al. LRRK2 variation and dementia with Lewy bodies. Parkinsonism Relat Disord. 2016;31:98–103.
Labbe C, Ogaki K, Lorenzo-Betancor O, Soto-Ortolaza AI, Walton RL, Rayaprolu S, et al. Role for the microtubule-associated protein tau variant p.A152T in risk of alpha-synucleinopathies. Neurology. 2015;85:1680–6.
McAleese KE, Walker L, Erskine D, Thomas AJ, McKeith IG, Attems J. TDP-43 pathology in Alzheimer’s disease, dementia with Lewy bodies and ageing. Brain Pathol. 2017;27:472–9.
Arai T, Mackenzie IR, Hasegawa M, Nonoka T, Niizato K, Tsuchiya K, et al. Phosphorylated TDP-43 in Alzheimer’s disease and dementia with Lewy bodies. Acta Neuropathol. 2009;117:125–36.
Nakashima-Yasuda H, Uryu K, Robinson J, Xie SX, Hurtig H, Duda JE, et al. Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 2007;114:221–9.
Uchino A, Takao M, Hatsuta H, Sumikura H, Nakano Y, Nogami A, et al. Incidence and extent of TDP-43 accumulation in aging human brain. Acta Neuropathol Commun. 2015;3:35.
Yokota O, Davidson Y, Arai T, Hasegawa M, Akiyama H, Ishizu H, et al. Effect of topographical distribution of alpha-synuclein pathology on TDP-43 accumulation in Lewy body disease. Acta Neuropathol. 2010;120:789–801.
McKee AC, Stern RA, Nowinski CJ, Stein TD, Alvarez VE, Daneshvar DH, et al. The spectrum of disease in chronic traumatic encephalopathy. Brain. 2013;136:43–64.
Atherton K, Han X, Chung J, Cherry JD, Baucom Z, Saltiel N, et al. Association of APOE genotypes and chronic traumatic encephalopathy. JAMA Neurol. 2022;79:787–96.
Cherry JD, Mez J, Crary JF, Tripodis Y, Alvarez VE, Mahar I, et al. Variation in TMEM106B in chronic traumatic encephalopathy. Acta Neuropathol Commun. 2018;6:115.
van Amerongen S, Kamps S, Kaijser KKM, Pijnenburg YAL, Scheltens P, Teunissen CE, et al. Severe CTE and TDP-43 pathology in a former professional soccer player with dementia: a clinicopathological case report and review of the literature. Acta Neuropathol Commun. 2023;11:77.
Arriagada PV, Growdon JH, Hedley-Whyte ET, Hyman BT. Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer’s disease. Neurology. 1992;42:631–9.
Gao J, Wang L, Huntley ML, Perry G, Wang X. Pathomechanisms of TDP-43 in neurodegeneration. J Neurochem 2018. https://doi.org/10.1111/jnc.14327.
Ayala YM, Pantano S, D’Ambrogio A, Buratti E, Brindisi A, Marchetti C, et al. Human, Drosophila, and C.elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol. 2005;348:575–88.
Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–808.
Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–43.
Prudencio M, Jansen-West KR, Lee WC, Gendron TF, Zhang YJ, Xu YF, et al. Misregulation of human sortilin splicing leads to the generation of a nonfunctional progranulin receptor. Proc Natl Acad Sci USA. 2012;109:21510–5.
Fiesel FC, Weber SS, Supper J, Zell A, Kahle PJ. TDP-43 regulates global translational yield by splicing of exon junction complex component SKAR. Nucleic Acids Res. 2012;40:2668–82.
Shiga A, Ishihara T, Miyashita A, Kuwabara M, Kato T, Watanabe N, et al. Alteration of POLDIP3 splicing associated with loss of function of TDP-43 in tissues affected with ALS. PLoS ONE. 2012;7:e43120.
Mohagheghi F, Prudencio M, Stuani C, Cook C, Jansen-West K, Dickson DW, et al. TDP-43 functions within a network of hnRNP proteins to inhibit the production of a truncated human SORT1 receptor. Hum Mol Genet. 2016;25:534–45.
De Conti L, Akinyi MV, Mendoza-Maldonado R, Romano M, Baralle M, Buratti E. TDP-43 affects splicing profiles and isoform production of genes involved in the apoptotic and mitotic cellular pathways. Nucleic Acids Res. 2015;43:8990–9005.
Ayala YM, De Conti L, Avendano-Vazquez SE, Dhir A, Romano M, D’Ambrogio A, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–88.
Colombrita C, Onesto E, Buratti E, de la Grange P, Gumina V, Baralle FE, et al. From transcriptomic to protein level changes in TDP-43 and FUS loss-of-function cell models. Biochim Biophys Acta. 2015;1849:1398–410.
Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–8.
Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–68.
Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349:650–5.
Klim JR, Williams LA, Limone F, Guerra San Juan I, Davis-Dusenbery BN, Mordes DA, et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat Neurosci. 2019;22:167–79.
Melamed Z, Lopez-Erauskin J, Baughn MW, Zhang O, Drenner K, Sun Y, et al. Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat Neurosci. 2019;22:180–90.
Brown AL, Wilkins OG, Keuss MJ, Hill SE, Zanovello M, Lee WC, et al. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature. 2022;603:131–7.
Ma XR, Prudencio M, Koike Y, Vatsavayai SC, Kim G, Harbinski F, et al. TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A. Nature. 2022;603:124–30.
Seddighi S, Qi YA, Brown AL, Wilkins OG, Bereda C, Belair C, et al. Mis-spliced transcripts generate de novo proteins in TDP-43-related ALS/FTD. Sci Transl Med. 2024;16:eadg7162.
Irwin KE, Jasin P, Braunstein KE, Sinha IR, Garret MA, Bowden KD, et al. A fluid biomarker reveals loss of TDP-43 splicing repression in presymptomatic ALS-FTD. Nat Med. 2024;30:382–93.
Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–5.
Susnjar U, Skrabar N, Brown AL, Abbassi Y, Phatnani H, Consortium NA. et al. Cell environment shapes TDP-43 function with implications in neuronal and muscle disease. Commun Biol. 2022;5:314.
Benard G, Bellance N, James D, Parrone P, Fernandez H, Letellier T, et al. Mitochondrial bioenergetics and structural network organization. J Cell Sci. 2007;120:838–48.
Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–10.
Kann O, Kovacs R. Mitochondria and neuronal activity. Am J Physiol. 2007;292:C641–657.
Johri A, Beal MF. Mitochondrial dysfunction in neurodegenerative diseases. J Pharmacol Exp Ther. 2012;342:619–30.
Xu YF, Gendron TF, Zhang YJ, Lin WL, D’Alton S, Sheng H, et al. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci. 2010;30:10851–9.
Wang W, Li L, Lin WL, Dickson DW, Petrucelli L, Zhang T, et al. The ALS disease-associated mutant TDP-43 impairs mitochondrial dynamics and function in motor neurons. Hum Mol Genet. 2013;22:4706–19.
Magrane J, Cortez C, Gan WB, Manfredi G. Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Human Mol Genet. 2014;23:1413–24.
Lu J, Duan W, Guo Y, Jiang H, Li Z, Huang J, et al. Mitochondrial dysfunction in human TDP-43 transfected NSC34 cell lines and the protective effect of dimethoxy curcumin. Brain Res Bull. 2012;89:185–90.
Stribl C, Samara A, Trumbach D, Peis R, Neumann M, Fuchs H, et al. Mitochondrial dysfunction and decrease in body weight of a transgenic knock-in mouse model for TDP-43. J Biol Chem. 2014;289:10769–84.
Salvatori I, Ferri A, Scaricamazza S, Giovannelli I, Serrano A, Rossi S, et al. Differential toxicity of TDP-43 isoforms depends on their sub-mitochondrial localization in neuronal cells. J Neurochem. 2018;146:585–97.
Genin EC, Bannwarth S, Lespinasse F, Ortega-Vila B, Fragaki K, Itoh K, et al. Loss of MICOS complex integrity and mitochondrial damage, but not TDP-43 mitochondrial localisation, are likely associated with severity of CHCHD10-related diseases. Neurobiol Dis. 2018;119:159–71.
Davis SA, Itaman S, Khalid-Janney CM, Sherard JA, Dowell JA, Cairns NJ, et al. TDP-43 interacts with mitochondrial proteins critical for mitophagy and mitochondrial dynamics. Neurosci Lett. 2018;678:8–15.
Woo JA, Liu T, Trotter C, Fang CC, De Narvaez E, LePochat P, et al. Loss of function CHCHD10 mutations in cytoplasmic TDP-43 accumulation and synaptic integrity. Nat Commun. 2017;8:15558.
Izumikawa K, Nobe Y, Yoshikawa H, Ishikawa H, Miura Y, Nakayama H, et al. TDP-43 stabilises the processing intermediates of mitochondrial transcripts. Sci Rep. 2017;7:7709.
Kawamata H, Peixoto P, Konrad C, Palomo G, Bredvik K, Gerges M, et al. Mutant TDP-43 does not impair mitochondrial bioenergetics in vitro and in vivo. Molecular Neurodegeneration. 2017;12:37.
Wang W, Wang L, Lu J, Siedlak SL, Fujioka H, Liang J, et al. The inhibition of TDP-43 mitochondrial localization blocks its neuronal toxicity. Nat Med. 2016;22:869–78.
Yu CH, Davidson S, Harapas CR, Hilton JB, Mlodzianoski MJ, Laohamonthonkul P, et al. TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell. 2020;183:636–49.e618.
Baek M, Choe YJ, Bannwarth S, Kim J, Maitra S, Dorn GW, et al. TDP-43 and PINK1 mediate CHCHD10(S59L) mutation-induced defects in Drosophila and in vitro. Nat Commun. 2021;12:1924.
Wang P, Deng J, Dong J, Liu J, Bigio EH, Mesulam M, et al. TDP-43 induces mitochondrial damage and activates the mitochondrial unfolded protein response. PLoS Genet. 2019;15:e1007947.
Zuo X, Zhou J, Li Y, Wu K, Chen Z, Luo Z, et al. TDP-43 aggregation induced by oxidative stress causes global mitochondrial imbalance in ALS. Nat Struct Mol Biol. 2021;28:132–42.
Stoica R, De Vos KJ, Paillusson S, Mueller S, Sancho RM, Lau KF, et al. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun. 2014;5:3996.
Ceron-Codorniu M, Torres P, Fernandez-Bernal A, Rico-Rios S, Serrano JC, Miralles MP, et al. TDP-43 dysfunction leads to bioenergetic failure and lipid metabolic rewiring in human cells. Redox Biol. 2024;75:103301.
Wang W, Arakawa H, Wang L, Okolo O, Siedlak SL, Jiang Y, et al. Motor-coordinative and cognitive dysfunction caused by mutant TDP-43 could be reversed by inhibiting its mitochondrial localization. Mol Ther. 2017;25:127–39.
Gao J, Wang L, Gao C, Arakawa H, Perry G, Wang X. TDP-43 inhibitory peptide alleviates neurodegeneration and memory loss in an APP transgenic mouse model for Alzheimer’s disease. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165580.
Das S, Vera M, Gandin V, Singer RH, Tutucci E. Intracellular mRNA transport and localized translation. Nat Rev Mol Cell Biol. 2021;22:483–504.
Russo A, Scardigli R, La Regina F, Murray ME, Romano N, Dickson DW, et al. Increased cytoplasmic TDP-43 reduces global protein synthesis by interacting with RACK1 on polyribosomes. Hum Mol Genet. 2017;26:1407–18.
MacNair L, Xiao S, Miletic D, Ghani M, Julien JP, Keith J, et al. MTHFSD and DDX58 are novel RNA-binding proteins abnormally regulated in amyotrophic lateral sclerosis. Brain. 2016;139:86–100.
Charif SE, Luchelli L, Vila A, Blaustein M, Igaz LM. Cytoplasmic expression of the ALS/FTD-related protein TDP-43 decreases global translation both in vitro and in vivo. Front Cell Neurosci. 2020;14:594561.
Lehmkuhl EM, Loganathan S, Alsop E, Blythe AD, Kovalik T, Mortimore NP, et al. TDP-43 proteinopathy alters the ribosome association of multiple mRNAs including the glypican Dally-like protein (Dlp)/GPC6. Acta Neuropathol Commun. 2021;9:52.
Altman T, Ionescu A, Ibraheem A, Priesmann D, Gradus-Pery T, Farberov L, et al. Axonal TDP-43 condensates drive neuromuscular junction disruption through inhibition of local synthesis of nuclear encoded mitochondrial proteins. Nat Commun. 2021;12:6914.
Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–20.
Chu JF, Majumder P, Chatterjee B, Huang SL, Shen CJ. TDP-43 regulates coupled dendritic mRNA transport-translation processes in co-operation with FMRP and Staufen1. Cell Rep. 2019;29:3118–33.e3116.
Majumder P, Chen YT, Bose JK, Wu CC, Cheng WC, Cheng SJ, et al. TDP-43 regulates the mammalian spinogenesis through translational repression of Rac1. Acta Neuropathol. 2012;124:231–45.
Nagano S, Jinno J, Abdelhamid RF, Jin Y, Shibata M, Watanabe S, et al. TDP-43 transports ribosomal protein mRNA to regulate axonal local translation in neuronal axons. Acta Neuropathol. 2020;140:695–713.
Briese M, Saal-Bauernschubert L, Luningschror P, Moradi M, Dombert B, Surrey V, et al. Loss of Tdp-43 disrupts the axonal transcriptome of motoneurons accompanied by impaired axonal translation and mitochondria function. Acta Neuropathol Commun. 2020;8:116.
Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58.
Ditlev JA, Case LB, Rosen MK. Who’s in and who’s out-compositional control of biomolecular condensates. J Mol Biol. 2018;430:4666–84.
Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis-Marks MR, Needham PG, et al. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron. 2019;102:321–38.e328.
Gasset-Rosa F, Lu S, Yu H, Chen C, Melamed Z, Guo L, et al. Cytoplasmic TDP-43 De-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron. 2019;102:339–57.e337.
Li HR, Chiang WC, Chou PC, Wang WJ, Huang JR. TAR DNA-binding protein 43 (TDP-43) liquid-liquid phase separation is mediated by just a few aromatic residues. J Biol Chem. 2018;293:6090–8.
Li HR, Chen TC, Hsiao CL, Shi L, Chou CY, Huang JR. The physical forces mediating self-association and phase-separation in the C-terminal domain of TDP-43. Biochim Biophys Acta Proteins Proteom. 2018;1866:214–23.
Lim LZ, Wei YY, Lu YM, Song JX. ALS-causing mutations significantly perturb the self-assembly and interaction with nucleic acid of the intrinsically disordered prion-like domain of TDP-43. Plos Biology. 2016;14:e1002338.
Conicella AE, Zerze GH, Mittal J, Fawzi NL. ALS mutations disrupt phase separation mediated by alpha-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24:1537–49.
Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–33.
Gao J, Wang L, Ren X, Dunn JR, Peters A, Miyagi M, et al. Translational regulation in the brain by TDP-43 phase separation. J Cell Biol. 2021;220:e202101019.
Dewey CM, Cenik B, Sephton CF, Dries DR, Mayer P 3rd, Good SK, et al. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol. 2011;31:1098–108.
Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, et al. TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem. 2009;111:1051–61.
Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderweyde T, Citro A, Mehta T, et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS ONE. 2010;5:e13250.
Wolozin B, Ivanov P. Stress granules and neurodegeneration. Nat Rev Neurosci. 2019;20:649–66.
Aulas A, Stabile S, Vande Velde C. Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol Neurodegener. 2012;7:54.
Mori F, Yasui H, Miki Y, Kon T, Arai A, Kurotaki H, et al. Colocalization of TDP-43 and stress granules at the early stage of TDP-43 aggregation in amyotrophic lateral sclerosis. Brain Pathol. 2024;34:e13215.
McGurk L, Gomes E, Guo L, Mojsilovic-Petrovic J, Tran V, Kalb RG, et al. Poly(ADP-Ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol Cell. 2018;71:703–17.e709.
Hasegawa M, Arai T, Nonaka T, Kametani F, Yoshida M, Hashizume Y, et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann Neurol. 2008;64:60–70.
Liachko NF, McMillan PJ, Guthrie CR, Bird TD, Leverenz JB, Kraemer BC. CDC7 inhibition blocks pathological TDP-43 phosphorylation and neurodegeneration. Ann Neurol. 2013;74:39–52.
Nonaka T, Suzuki G, Tanaka Y, Kametani F, Hirai S, Okado H, et al. Phosphorylation of TAR DNA-binding protein of 43 kDa (TDP-43) by truncated casein kinase 1delta triggers mislocalization and accumulation of TDP-43. J Biol Chem. 2016;291:5473–83.
Hicks DA, Cross LL, Williamson R, Rattray M. Endoplasmic reticulum stress signalling induces casein kinase 1-dependent formation of cytosolic TDP-43 inclusions in motor neuron-like cells. Neurochem Res. 2020;45:1354–64.
Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 2018;37:e97452.
Cohen TJ, Hwang AW, Unger T, Trojanowski JQ, Lee VM. Redox signalling directly regulates TDP-43 via cysteine oxidation and disulphide cross-linking. EMBO J. 2012;31:1241–52.
Verde EM, Antoniani F, Mediani L, Secco V, Crotti S, Ferrara MC, et al. SUMO2/3 conjugation of TDP-43 protects against aggregation. Sci Adv. 2025;11:eadq2475.
Maraschi A, Gumina V, Dragotto J, Colombrita C, Mompean M, Buratti E, et al. SUMOylation regulates TDP-43 splicing activity and nucleocytoplasmic distribution. Mol Neurobiol. 2021;58:5682–702.
Marino R, Buccarello L, Hassanzadeh K, Akhtari K, Palaniappan S, Corbo M, et al. A novel cell-permeable peptide prevents protein SUMOylation and supports the mislocalization and aggregation of TDP-43. Neurobiol Dis. 2023;188:106342.
Xiao S, Sanelli T, Chiang H, Sun Y, Chakrabartty A, Keith J, et al. Low molecular weight species of TDP-43 generated by abnormal splicing form inclusions in amyotrophic lateral sclerosis and result in motor neuron death. Acta Neuropathol. 2015;130:49–61.
Zhang YJ, Xu YF, Dickey CA, Buratti E, Baralle F, Bailey R, et al. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci. 2007;27:10530–4.
Rutherford NJ, Zhang YJ, Baker M, Gass JM, Finch NA, Xu YF, et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008;4:e1000193.
Zhang YJ, Xu YF, Cook C, Gendron TF, Roettges P, Link CD, et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci USA. 2009;106:7607–12.
Yamashita T, Hideyama T, Hachiga K, Teramoto S, Takano J, Iwata N, et al. A role for calpain-dependent cleavage of TDP-43 in amyotrophic lateral sclerosis pathology. Nat Commun. 2012;3:1307.
Suzuki H, Lee K, Matsuoka M. TDP-43-induced death is associated with altered regulation of BIM and Bcl-xL and attenuated by caspase-mediated TDP-43 cleavage. J Biol Chem. 2011;286:13171–83.
Igaz LM, Kwong LK, Chen-Plotkin A, Winton MJ, Unger TL, Xu Y, et al. Expression of TDP-43 C-terminal Fragments in Vitro Recapitulates Pathological Features of TDP-43 Proteinopathies. J Biol Chem. 2009;284:8516–24.
Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–39.
Fuentealba RA, Udan M, Bell S, Wegorzewska I, Shao J, Diamond MI, et al. Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J Biol Chem. 2010;285:26304–14.
D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–66.
Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature. 2018;555:475–82.
Chou CC, Zhang Y, Umoh ME, Vaughan SW, Lorenzini I, Liu F, et al. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat Neurosci. 2018;21:228–39.
Baskerville V, Rapuri S, Mehlhop E, Coyne AN. SUN1 facilitates CHMP7 nuclear influx and injury cascades in sporadic amyotrophic lateral sclerosis. Brain. 2024;147:109–21.
Coyne AN, Zaepfel BL, Hayes L, Fitchman B, Salzberg Y, Luo EC, et al. G(4)C(2) repeat RNA Initiates a POM121-mediated reduction in specific nucleoporins in C9orf72 ALS/FTD. Neuron. 2020;107:1124–40.e1111.
Coyne AN, Baskerville V, Zaepfel BL, Dickson DW, Rigo F, Bennett F, et al. Nuclear accumulation of CHMP7 initiates nuclear pore complex injury and subsequent TDP-43 dysfunction in sporadic and familial ALS. Sci Transl Med. 2021;13:eabe1923.
Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–33.
Giampetruzzi A, Danielson EW, Gumina V, Jeon M, Boopathy S, Brown RH, et al. Modulation of actin polymerization affects nucleocytoplasmic transport in multiple forms of amyotrophic lateral sclerosis. Nat Commun. 2019;10:3827.
Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–9.
Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61.
Miyagi T, Kanekura K. Intricate roles of spacers and stickers of Arg-rich C9ORF72 dipeptide repeat proteins; from toxicity to targeting to membraneless organelles. Organelle 2024; 1. https://doi.org/10.61747/0ifp.202311001.
Khalil B, Chhangani D, Wren MC, Smith CL, Lee JH, Li X, et al. Nuclear import receptors are recruited by FG-nucleoporins to rescue hallmarks of TDP-43 proteinopathy. Mol Neurodegener. 2022;17:80.
Takeuchi T, Maeta K, Ding X, Oe Y, Takeda A, Inoue M, et al. Sustained therapeutic benefits by transient reduction of TDP-43 using ENA-modified antisense oligonucleotides in ALS/FTD mice. Mol Ther Nucleic Acids. 2023;31:353–66.
Baughn MW, Melamed Z, Lopez-Erauskin J, Beccari MS, Ling K, Zuberi A, et al. Mechanism of STMN2 cryptic splice-polyadenylation and its correction for TDP-43 proteinopathies. Science. 2023;379:1140–9.
Wilkins OG, Chien M, Wlaschin JJ, Barattucci S, Harley P, Mattedi F, et al. Creation of de novo cryptic splicing for ALS and FTD precision medicine. Science. 2024;386:61–69.
Matthew JKPH, Eugeni R, Rachel EJ, Matteo Z, Oscar GW. Loss of TDP-43 induces synaptic dysfunction that is rescued by UNC13A splice-switching ASOs. bioRxiv. 2024.
Prasad A, Raju G, Sivalingam V, Girdhar A, Verma M, Vats A, et al. An acridine derivative, [4,5-bis(N-carboxy methyl imidazolium)methylacridine] dibromide, shows anti-TDP-43 aggregation effect in ALS disease models. Sci Rep. 2016;6:39490.
Babinchak WM, Dumm BK, Venus S, Boyko S, Putnam AA, Jankowsky E, et al. Small molecules as potent biphasic modulators of protein liquid-liquid phase separation. Nat Commun. 2020;11:5574.
Francois-Moutal L, Felemban R, Scott DD, Sayegh MR, Miranda VG, Perez-Miller S, et al. Small molecule targeting TDP-43’s RNA recognition motifs reduces locomotor defects in a drosophila model of amyotrophic lateral sclerosis (ALS). ACS Chem Biol. 2019;14:2006–13.
Mollasalehi N, Francois-Moutal L, Scott DD, Tello JA, Williams H, Mahoney B, et al. An allosteric modulator of RNA binding targeting the N-terminal domain of TDP-43 yields neuroprotective properties. ACS Chem Biol. 2020;15:2854–9.
Tseng YL, Lu PC, Lee CC, He RY, Huang YA, Tseng YC, et al. Degradation of neurodegenerative disease-associated TDP-43 aggregates and oligomers via a proteolysis-targeting chimera. J Biomed Sci. 2023;30:27.
Author information
Authors and Affiliations
Contributions
JL and XW provided expert insights, critical revisions, and conceptual guidance. XW finalized the manuscript. JG, MD, SQ, DS, LV, JL, and XW all contributed to the literature review and drafting of the manuscript. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gao, J., Ding, M., Qin, S. et al. The mechanisms underlying TDP-43-associated neurodegeneration in Alzheimer’s disease and related dementias. Mol Psychiatry (2025). https://doi.org/10.1038/s41380-025-03089-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-025-03089-8