Abstract
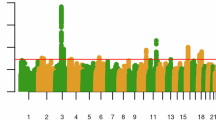
Cannabis is one of the most widely used drugs globally. We performed genome-wide association studies (GWASs) of lifetime (N = 131,895) and frequency (N = 73,374) of cannabis use. For lifetime cannabis use, we identified two loci, one near CADM2 (rs35827242, p = 4.63E-12) and another near GRM3 (rs12673181, p = 6.90E-09). For frequency of cannabis use, we identified one locus near CADM2 (rs4856591, p = 8.10E-09; r2 = 0.76 with rs35827242). Lifetime and frequency of cannabis use were heritable (12.88 vs. 6.63%) and genetically correlated with previous GWASs of lifetime use and cannabis use disorder (CUD), as well as other substance use and cognitive traits. Polygenic scores (PGSs) for lifetime and frequency of cannabis use predicted cannabis use phenotypes in All of Us participants. A phenome-wide association study using a PGS for lifetime cannabis use to interrogate a hospital cohort replicated prior associations with substance use and mood disorders, and uncovered novel associations with celiac and infectious diseases. This work demonstrates the utility of pre-addiction phenotypes in cannabis use genomic discovery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
We provide 23andMe summary statistics for the top 10,000 independent SNPs. 23andMe GWAS summary statistics will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please visit https://research.23andme.com/collaborate/#dataset-access/ for more information and to apply to access the data. We will share the Jupyter notebooks used for PGS analysis in AoU with registered All of Us researchers upon request.
References
United Nations Office on Drug and Crime. Booklet 3 - drug market trends of cannabis and opioids. Vienna; 2022.
Pacek LR, Mauro PM, Martins SS. Perceived risk of regular cannabis use in the United States from 2002 to 2012: differences by sex, age, and race/ethnicity. Drug Alcohol Depend. 2015;149:232–44.
Statistics Canada. Mental and substance use disorders in Canada. 2013. (Catalogue no.82-624-X).
Rotermann M. Looking back from 2020, how cannabis use and related behaviours changed in Canada. Health Rep. 2021;32:3–14.
Whiting PF, Wolff RF, Deshpande S, Di Nisio M, Duffy S, Hernandez AV, et al. Cannabinoids for medical use: a systematic review and meta-analysis. JAMA. 2015;313:2456–73.
Bellocchio L, Inchingolo AD, Inchingolo AM, Lorusso F, Malcangi G, Santacroce L, et al. Cannabinoids drugs and oral health-from recreational side-effects to medicinal purposes: a systematic review. Int J Mol Sci. 2021;22:8329.
Diep C, Tian C, Vachhani K, Won C, Wijeysundera DN, Clarke H, et al. Recent cannabis use and nightly sleep duration in adults: a population analysis of the NHANES from 2005 to 2018. Reg Anesth Pain Med. 2022;47:100–4.
Hasin DS, Kerridge BT, Saha TD, Huang B, Pickering R, Smith SM, et al. Prevalence and correlates of DSM-5 cannabis use disorder, 2012–2013: findings from the national epidemiologic survey on alcohol and related Conditions-III. Am J Psychiatry. 2016;173:588–99.
Hayley AC, Stough C, Downey LA. DSM-5 cannabis use disorder, substance use and DSM-5 specific substance-use disorders: evaluating comorbidity in a population-based sample. Eur Neuropsychopharmacol. 2017;27:732–43.
Howard J, Osborne J. Cannabis and work: need for more research. Am J Ind Med. 2020;63:963–72.
Keen L 2nd, Turner AD, George L, Lawrence K. Cannabis use disorder severity and sleep quality among undergraduates attending a Historically Black University. Addict Behav. 2022;134:107414.
Lo JO, Hedges JC, Girardi G. Impact of cannabinoids on pregnancy, reproductive health, and offspring outcomes. Am J Obstet Gynecol. 2022;227:571–81.
Pacek LR, Herrmann ES, Smith MT, Vandrey R. Sleep continuity, architecture and quality among treatment-seeking cannabis users: an in-home, unattended polysomnographic study. Exp Clin Psychopharmacol. 2017;25:295–302.
Page RL 2nd, Allen LA, Kloner RA, Carriker CR, Martel C, et al. Medical marijuana, recreational cannabis, and cardiovascular health: a scientific statement from the American Heart Association. Circulation. 2020;142:e131–52.
Feingold D, Livne O, Rehm J, Lev-Ran S. Probability and correlates of transition from cannabis use to DSM-5 cannabis use disorder: results from a large-scale nationally representative study. Drug Alcohol Rev. 2020;39:142–51.
American Psychiatric Association. Substance-related and addictive disorders. Diagnostic and statistical manual of mental disorders, 5th edn. Washington, D.C.; 2013. https://doi.org/10.1176/appi.books.9780890425596.dsm16.
Hines LA, Morley KI, Rijsdijk F, Strang J, Agrawal A, Nelson EC, et al. Overlap of heritable influences between cannabis use disorder, frequency of use and opportunity to use cannabis: trivariate twin modelling and implications for genetic design. Psychol Med. 2018;48:2786–93.
Kendler KS, Ohlsson H, Maes HH, Sundquist K, Lichtenstein P, Sundquist J. A population-based Swedish Twin and Sibling Study of cannabis, stimulant and sedative abuse in men. Drug Alcohol Depend. 2015;149:49–54.
Verweij KJ, Zietsch BP, Lynskey MT, Medland SE, Neale MC, Martin NG, et al. Genetic and environmental influences on cannabis use initiation and problematic use: a meta-analysis of twin studies. Addiction. 2010;105:417–30.
Demontis D, Rajagopal VM, Thorgeirsson TE, Als TD, Grove J, Leppala K, et al. Genome-wide association study implicates CHRNA2 in cannabis use disorder. Nat Neurosci. 2019;22:1066–74.
Johnson EC, Demontis D, Thorgeirsson TE, Walters RK, Polimanti R, Hatoum AS, et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry. 2020;7:1032–45.
Levey DF, Galimberti M, Deak JD, Wendt FR, Bhattacharya A, Koller D, et al. Multi-ancestry genome-wide association study of cannabis use disorder yields insight into disease biology and public health implications. Nat Genet. 2023;55:2094–103.
Xu H, Toikumo S, Crist RC, Glogowska K, Jinwala Z, Deak JD, et al. Identifying genetic loci and phenomic associations of substance use traits: a multi-trait analysis of GWAS (MTAG) study. Addiction. 2023;118:1942–52.
Sanchez-Roige S, Palmer AA, Clarke TK. Recent efforts to dissect the genetic basis of alcohol use and abuse. Biol Psychiatry. 2020;87:609–18.
McLellan AT, Koob GF, Volkow ND. Preaddiction-A missing concept for treating substance use disorders. JAMA Psychiatry. 2022;79:749–51.
Agrawal A, Madden PA, Bucholz KK, Heath AC, Lynskey MT. Initial reactions to tobacco and cannabis smoking: a Twin study. Addiction. 2014;109:663–71.
Degenhardt L, Coffey C, Carlin JB, Swift W, Moore E, Patton GC. Outcomes of occasional cannabis use in adolescence: 10-year follow-up study in Victoria, Australia. Br J Psychiatry. 2010;196:290–5.
Scherrer JF, Grant JD, Duncan AE, Sartor CE, Haber JR, Jacob T, et al. Subjective effects to cannabis are associated with use, abuse and dependence after adjusting for genetic and environmental influences. Drug Alcohol Depend. 2009;105:76–82.
Swift W, Coffey C, Carlin JB, Degenhardt L, Patton GC. Adolescent cannabis users at 24 years: trajectories to regular weekly use and dependence in young adulthood. Addiction. 2008;103:1361–70.
Windle M, Wiesner M. Trajectories of marijuana use from adolescence to young adulthood: predictors and outcomes. Dev Psychopathol. 2004;16:1007–27.
Lyons MJ, Toomey R, Meyer JM, Green AI, Eisen SA, Goldberg J, et al. How do genes influence marijuana use? The role of subjective effects. Addiction. 1997;92:409–17.
Leung J, Chan GCK, Hides L, Hall WD. What is the prevalence and risk of cannabis use disorders among people who use cannabis? A systematic review and meta-analysis. Addict Behav. 2020;109:106479.
Haberstick BC, Zeiger JS, Corley RP, Hopfer CJ, Stallings MC, Rhee SH, et al. Common and drug-specific genetic influences on subjective effects to alcohol, tobacco and marijuana use. Addiction. 2011;106:215–24.
Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL, et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci. 2018;21:1161–70.
Stringer S, Minica CC, Verweij KJ, Mbarek H, Bernard M, Derringer J, et al. Genome-wide association study of lifetime cannabis use based on a large meta-analytic sample of 32 330 subjects from the International Cannabis Consortium. Transl Psychiatry. 2016;6:e769.
Sanchez-Roige S, Palmer AA. Emerging phenotyping strategies will advance our understanding of psychiatric genetics. Nat Neurosci. 2020;23:475–80.
Thorpe HHA, Talhat MA, Khokhar JY. High genes: genetic underpinnings of cannabis use phenotypes. Prog Neuropsychopharmacol Biol Psychiatry. 2021;106:110164.
McBain RK, Wong EC, Breslau J, Shearer AL, Cefalu MS, Roth E, et al. State medical marijuana laws, cannabis use and cannabis use disorder among adults with elevated psychological distress. Drug Alcohol Depend. 2020;215:108191.
Hughes JR, Naud S, Budney AJ, Fingar JR, Callas PW. Attempts to stop or reduce daily cannabis use: an Intensive Natural History study. Psychol Addict Behav. 2016;30:389–97.
Mallard TT, Sanchez-Roige S. Dimensional phenotypes in psychiatric genetics: lessons from genome-wide association studies of alcohol use phenotypes. Complex Psychiatry. 2021;7:45–8.
Toikumo S, Jennings MV, Pham BK, Lee H, Mallard TT, Bianchi SB, et al. Multi-ancestry meta-analysis of tobacco use disorder identifies 461 potential risk genes and reveals associations with multiple health outcomes. Nat Human Behav. 2024;8:1177–93.
Sanchez-Roige S, Jennings MV, Thorpe HHA, Mallari JE, van der Werf LC, Bianchi SB, et al. CADM2 is implicated in impulsive personality and numerous other traits by genome- and phenome-wide association studies in humans and mice. Transl Psychiatry. 2023;13:167.
Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am J Hum Genet. 2015;96:37–53.
National Academies of Sciences, Engineering, and Medicine Health and Medicine Division, Division of Behavioral and Social Sciences and Education, Board on Health Sciences Policy, Committee on population, committee on the use of race, ethnicity, and ancestry as population descriptors in genomics research. Using population descriptors in genetics and genomics research: a new framework for an evolving field. Washington (DC): National Academies Press (US); 2023.
Durand EY, Do CB, Mountain JL, Macpherson JM Ancestry composition: a novel, efficient pipeline for ancestry deconvolution. bioRxiv [Preprint]. 2014.
Hatoum AS, Colbert SMC, Johnson EC, Huggett SB, Deak JD, Pathak G, et al. Multivariate genome-wide association meta-analysis of over 1 million subjects identifies loci underlying multiple substance use disorders. Nat Ment Health. 2023;1:210–23.
Yin B, Wang X, Huang T, Jia J. Shared genetics and causality between decaffeinated coffee consumption and neuropsychiatric diseases: a large-scale genome-wide cross-trait analysis and mendelian randomization analysis. Front Psychiatry. 2022;13:910432.
Agrawal A, Chou YL, Carey CE, Baranger DAA, Zhang B, Sherva R, et al. Genome-wide association study identifies a novel locus for cannabis dependence. Mol Psychiatry. 2018;23:1293–302.
Sherva R, Wang Q, Kranzler H, Zhao H, Koesterer R, Herman A, et al. Genome-wide Association Study of cannabis dependence severity, novel risk variants, and shared genetic risks. JAMA Psychiatry. 2016;73:472–80.
Agrawal A, Lynskey MT, Hinrichs A, Grucza R, Saccone SF, Krueger R, et al. A genome-wide association study of DSM-IV cannabis dependence. Addict Biol. 2011;16:514–8.
Minica CC, Dolan CV, Hottenga JJ, Pool R, Genome of the Netherlands Consortium, Fedko IO, et al. Heritability, SNP- and gene-based analyses of cannabis use initiation and age at onset. Behav Genet. 2015;45:503–13.
Zhao Y, Han X, Zheng ZL. Analysis of the brain transcriptome for substance-associated genes: An update on large-scale genome-wide association studies. Addict Biol. 2023;28:e13332.
Greco LA, Reay WR, Dayas CV, Cairns MJ. Exploring opportunities for drug repurposing and precision medicine in cannabis use disorder using genetics. Addict Biol. 2023;28:e13313.
Carreras-Gallo N, Dwaraka VB, Caceres A, Smith R, Mendez TL, Went H, et al. Impact of tobacco, alcohol, and marijuana on genome-wide DNA methylation and its relationship with hypertension. Epigenetics. 2023;18:2214392.
Cheng W, Parker N, Karadag N, Koch E, Hindley G, Icick R, et al. The relationship between cannabis use, schizophrenia, and bipolar disorder: a genetically informed study. Lancet Psychiatry. 2023;10:441–51.
Greco LA, Reay WR, Dayas CV, Cairns MJ. Pairwise genetic meta-analyses between schizophrenia and substance dependence phenotypes reveals novel association signals with pharmacological significance. Transl Psychiatry. 2022;12:403.
Minica CC, Verweij KJH, van der Most PJ, Mbarek H, Bernard M, van Eijk KR, et al. Genome-wide association meta-analysis of age at first cannabis use. Addiction. 2018;113:2073–86.
Sey NYA, Hu B, Mah W, Fauni H, McAfee JC, Rajarajan P, et al. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat Neurosci. 2020;23:583–93.
Barbeira, Dickinson AN, Bonazzola SP, Zheng R, Wheeler HE J, Torres JM, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9:1825.
Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5.
All of Us Research Program Genomics Investigators. Genomic data in the All of Us research program. Nature. 2024;627:340–6.
All of Us Research Program Investigators, Denny JC, Rutter JL, Goldstein DB, Philippakis A, Smoller JW, et al. The “All of Us” research program. N Engl J Med. 2019;381:668–76.
Ge T, Chen CY, Ni Y, Feng YA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10:1776.
Lee SH, Goddard ME, Wray NR, Visscher PM. A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012;36:214–24.
Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–9.
Dennis J, Sealock J, Levinson RT, Farber-Eger E, Franco J, Fong S, et al. Genetic risk for major depressive disorder and loneliness in sex-specific associations with coronary artery disease. Mol Psychiatry. 2021;26:4254–64.
Fogel AI, Akins MR, Krupp AJ, Stagi M, Stein V, Biederer T. SynCAMs organize synapses through heterophilic adhesion. J Neurosci. 2007;27:12516–30.
Yan X, Wang Z, Schmidt V, Gauert A, Willnow TE, Heinig M, et al. Cadm2 regulates body weight and energy homeostasis in mice. Mol Metab. 2018;8:180–8.
Tyler RE, Besheer J, Joffe ME. Advances in translating mGlu(2) and mGlu(3) receptor selective allosteric modulators as breakthrough treatments for affective disorders and alcohol use disorder. Pharmacol Biochem Behav. 2022;219:173450.
Karlsson Linner R, Mallard TT, Barr PB, Sanchez-Roige S, Madole JW, Driver MN, et al. Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat Neurosci. 2021;24:1367–76.
Agrawal A, Budney AJ, Lynskey MT. The co-occurring use and misuse of cannabis and tobacco: a review. Addiction. 2012;107:1221–33.
Galimberti M, Levey DF, Deak JD, Zhou H, Stein MB, Gelernter J. Genetic influences and causal pathways shared between cannabis use disorder and other substance use traits. Mol Psychiatry. 2024.
Mallard TT, Savage JE, Johnson EC, Huang Y, Edwards AC, Hottenga JJ, et al. Item-Level Genome-Wide Association Study of the alcohol use disorders identification test in three population-based cohorts. Am J Psychiatry. 2022;179:58–70.
Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun. 2019;10:1499.
Zhou H, Kember RL, Deak JD, Xu H, Toikumo S, Yuan K, et al. Multi-ancestry study of the genetics of problematic alcohol use in over 1 million individuals. Nat Med. 2023;29:3184–92.
Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, 23andMe Research Team, Substance Use Disorder Working Group of the Psychiatric Genomics Consortium. et al. Genome-Wide Association Study meta-analysis of the alcohol use disorders identification test (AUDIT) in two population-based cohorts. Am J Psychiatry. 2019;176:107–18.
Savage JE, Barr PB, Phung T, Lee YH, Zhang Y, Ge T, et al. Genetic heterogeneity across dimensions of alcohol use behaviors. medRxiv. 2023.2012.2026.23300537. 2023.
Saunders GRB, Wang X, Chen F, Jang SK, Liu M, Wang C, et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature. 2022;612:720–4.
Trubetskoy V, Pardinas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–8.
Lam M, Chen CY, Li Z, Martin AR, Bryois J, Ma X, et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet. 2019;51:1670–8.
Pardinas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–9.
Li Z, Chen J, Yu H, He L, Xu Y, Zhang D, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49:1576–83.
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7.
Nagel M, Watanabe K, Stringer S, Posthuma D, van der Sluis S. Item-level analyses reveal genetic heterogeneity in neuroticism. Nat Commun. 2018;9:905.
Luciano M, Hagenaars SP, Davies G, Hill WD, Clarke TK, Shirali M, et al. Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nat Genet. 2018;50:6–11.
Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–21.
Hysi PG, Choquet H, Khawaja AP, Wojciechowski R, Tedja MS, Yin J, et al. Meta-analysis of 542,934 subjects of European ancestry identifies new genes and mechanisms predisposing to refractive error and myopia. Nat Genet. 2020;52:401–7.
Tedja MS, Wojciechowski R, Hysi PG, Eriksson N, Furlotte NA, Verhoeven VJM, et al. Genome-wide association meta-analysis highlights light-induced signaling as a driver for refractive error. Nat Genet. 2018;50:834–48.
Watanabe K, Jansen PR, Savage JE, Nandakumar P, Wang X, 23andMe Research Team. et al. Genome-wide meta-analysis of insomnia prioritizes genes associated with metabolic and psychiatric pathways. Nat Genet. 2022;54:1125–32.
Di Menna L, Joffe ME, Iacovelli L, Orlando R, Lindsley CW, Mairesse J, et al. Functional partnership between mGlu3 and mGlu5 metabotropic glutamate receptors in the central nervous system. Neuropharmacology. 2018;128:301–13.
Ibrahim KS, Abd-Elrahman KS, El Mestikawy S, Ferguson SSG. Targeting vesicular glutamate transporter machinery: implications on metabotropic glutamate receptor 5 signaling and behavior. Mol Pharmacol. 2020;98:314–27.
Jung KM, Mangieri R, Stapleton C, Kim J, Fegley D, Wallace M, et al. Stimulation of endocannabinoid formation in brain slice cultures through activation of group I metabotropic glutamate receptors. Mol Pharmacol. 2005;68:1196–202.
GTEX portal. https://www.gtexportal.org/home/. Accessed 2024.
Schoeler T, Speed D, Porcu E, Pirastu N, Pingault JB, Kutalik Z. Participation bias in the UK Biobank distorts genetic associations and downstream analyses. Nat Hum Behav. 2023;7:1216–27.
Deak JD, Levey DF, Wendt FR, Zhou H, Galimberti M, Kranzler HR, et al. Genome-wide investigation of maximum habitual alcohol intake in us veterans in relation to alcohol consumption traits and alcohol use disorder. JAMA Netw Open. 2022;5:e2238880.
Pasman JA, Demange PA, Guloksuz S, Willemsen AHM, Abdellaoui A, Ten Have M, et al. Genetic risk for smoking: disentangling interplay between genes and socioeconomic status. Behav Genet. 2022;52:92–107.
Xu K, Li B, McGinnis KA, Vickers-Smith R, Dao C, Sun N, et al. Genome-wide association study of smoking trajectory and meta-analysis of smoking status in 842,000 individuals. Nat Commun. 2020;11:5302.
Zhou H, Sealock JM, Sanchez-Roige S, Clarke TK, Levey DF, Cheng Z, et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci. 2020;23:809–18.
Cai N, Revez JA, Adams MJ, Andlauer TFM, Breen G, Byrne EM, et al. Minimal phenotyping yields genome-wide association signals of low specificity for major depression. Nat Genet. 2020;52:437–47.
Evangelou E, Gao H, Chu C, Ntritsos G, Blakeley P, Butts AR, et al. New alcohol-related genes suggest shared genetic mechanisms with neuropsychiatric disorders. Nat Hum Behav. 2019;3:950–61.
Zhong VW, Kuang A, Danning RD, Kraft P, van Dam RM, Chasman DI, et al. A genome-wide association study of bitter and sweet beverage consumption. Hum Mol Genet. 2019;28:2449–57.
Sanchez-Roige S, Fontanillas P, Elson SL, Gray JC, de Wit H, MacKillop J, et al. Genome-wide association studies of impulsive personality traits (BIS-11 and UPPS-P) and drug experimentation in up to 22,861 adult research participants identify loci in the CACNA1I and CADM2 genes. J Neurosci. 2019;39:2562–72.
Karlsson Linner R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51:245–57.
Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–44.
Erzurumluoglu AM, Liu M, Jackson VE, Barnes DR, Datta G, Melbourne CA, et al. Meta-analysis of up to 622,409 individuals identifies 40 novel smoking behaviour associated genetic loci. Mol Psychiatry. 2020;25:2392–409.
Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019;104:65–75.
Clifton EAD, Perry JRB, Imamura F, Lotta LA, Brage S, Forouhi NG, et al. Genome-wide association study for risk taking propensity indicates shared pathways with body mass index. Commun Biol. 2018;1:36.
Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, et al. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N = 112 117). Mol Psychiatry. 2017;22:1376–84.
Brazel DM, Jiang Y, Hughey JM, Turcot V, Zhan X, Gong J, et al. Exome chip meta-analysis fine maps causal variants and elucidates the genetic architecture of rare coding variants in smoking and alcohol use. Biol Psychiatry. 2019;85:946–55.
Baselmans B, Hammerschlag AR, Noordijk S, Ip H, van der Zee M, de Geus E, et al. The genetic and neural substrates of externalizing behavior. Biol Psychiatry Glob Open Sci. 2022;2:389–99.
Strawbridge RJ, Ward J, Cullen B, Tunbridge EM, Hartz S, Bierut L, et al. Genome-wide analysis of self-reported risk-taking behaviour and cross-disorder genetic correlations in the UK Biobank cohort. Transl Psychiatry. 2018;8:39.
Mills MC, Tropf FC, Brazel DM, van Zuydam N, Vaez A, eQTLGen Consortium. et al. Identification of 371 genetic variants for age at first sex and birth linked to externalising behaviour. Nat Hum Behav. 2021;5:1717–30.
Johnson EC, Sanchez-Roige S, Acion L, Adams MJ, Bucholz KK, Chan G, et al. Polygenic contributions to alcohol use and alcohol use disorders across population-based and clinically ascertained samples. Psychol Med. 2021;51:1147–56.
Sun Y, Chang S, Wang F, Sun H, Ni Z, Yue W, et al. Genome-wide association study of alcohol dependence in male Han Chinese and cross-ethnic polygenic risk score comparison. Transl Psychiatry. 2019;9:249.
Lai D, Wetherill L, Bertelsen S, Carey CE, Kamarajan C, Kapoor M, et al. Genome-wide association studies of alcohol dependence, DSM-IV criterion count and individual criteria. Genes Brain Behav. 2019;18:e12579.
Chang LH, Couvy-Duchesne B, Liu M, Medland SE, Verhulst B, Benotsch EG, et al. Association between polygenic risk for tobacco or alcohol consumption and liability to licit and illicit substance use in young Australian adults. Drug Alcohol Depend. 2019;197:271–9.
Allegrini AG, Verweij KJH, Abdellaoui A, Treur JL, Hottenga JJ, Willemsen G, et al. Genetic vulnerability for smoking and cannabis use: associations with E-cigarette and water pipe use. Nicotine Tob Res. 2019;21:723–30.
Hodgson K, Coleman JRI, Hagenaars SP, Purves KL, Glanville K, Choi SW, et al. Cannabis use, depression and self-harm: phenotypic and genetic relationships. Addiction. 2020;115:482–92.
Smeland OB, Andreassen OA. Polygenic risk scores in psychiatry - large potential but still limited clinical utility. Eur Neuropsychopharmacol. 2021;51:68–70.
Kember RL, Hartwell EE, Xu H, Rotenberg J, Almasy L, Zhou H, et al. Phenome-wide association analysis of substance use disorders in a deeply phenotyped sample. Biol Psychiatry. 2023;93:536–45.
Johnson EC, Hatoum AS, Deak JD, Polimanti R, Murray RM, Edenberg HJ, et al. The relationship between cannabis and schizophrenia: a genetically informed perspective. Addiction. 2021;116:3227–34.
Reginsson GW, Ingason A, Euesden J, Bjornsdottir G, Olafsson S, Sigurdsson E, et al. Polygenic risk scores for schizophrenia and bipolar disorder associate with addiction. Addict Biol. 2018;23:485–92.
Johnson EC, Austin-Zimmerman I, Thorpe HHA, Levey DF, Baranger DAA, Colbert SMC et al. Cross-ancestry genetic investigation of schizophrenia, cannabis use disorder, and tobacco smoking. Neuropsychopharmacology. 2024;49:1655–65.
Hamilton I, Monaghan M. Cannabis and psychosis: are we any closer to understanding the relationship? Curr Psychiatry Rep. 2019;21:48.
Di Forti M, Quattrone D, Freeman TP, Tripoli G, Gayer-Anderson C, Quigley H, et al. The contribution of cannabis use to variation in the incidence of psychotic disorder across Europe (EU-GEI): a multicentre case-control study. Lancet Psychiatry. 2019;6:427–36.
Oluwoye O, Monroe-DeVita M, Burduli E, Chwastiak L, McPherson S, McClellan JM, et al. Impact of tobacco, alcohol and cannabis use on treatment outcomes among patients experiencing first episode psychosis: data from the national RAISE-ETP study. Early Interv Psychiatry. 2019;13:142–6.
Schoeler T, Monk A, Sami MB, Klamerus E, Foglia E, Brown R, et al. Continued versus discontinued cannabis use in patients with psychosis: a systematic review and meta-analysis. Lancet Psychiatry. 2016;3:215–25.
Durvasula A, Price AL Distinct explanations underlie gene-environment interactions in the UK Biobank. medRxiv 2023.
Abdellaoui A, Dolan CV, Verweij KJH, Nivard MG. Gene-environment correlations across geographic regions affect genome-wide association studies. Nat Genet. 2022;54:1345–54.
Thorpe HHA, Fontanillas P, Pham BK, Meredith JJ, Jennings MV, Courchesne-Krak NS, et al. Genome-wide association studies of coffee intake in UK/US participants of European ancestry uncover cohort-specific genetic associations. Neuropsychopharmacology. 2024;49:1609–18.
Jang SK, Saunders G, Liu M, andMe Research T, Jiang Y, Liu DJ, et al. Genetic correlation, pleiotropy, and causal associations between substance use and psychiatric disorder. Psychol Med. 2022;52:968–78.
Chang LH, Ong JS, An J, Verweij KJH, Vink JM, Pasman J, et al. Investigating the genetic and causal relationship between initiation or use of alcohol, caffeine, cannabis and nicotine. Drug Alcohol Depend. 2020;210:107966.
Abdellaoui A, Smit DJA, van den Brink W, Denys D, Verweij KJH. Genomic relationships across psychiatric disorders including substance use disorders. Drug Alcohol Depend. 2021;220:108535.
Waldman ID, Poore HE, Luningham JM, Yang J. Testing structural models of psychopathology at the genomic level. World Psychiatry. 2020;19:350–9.
Poore HE, Hatoum A, Mallard TT, Sanchez-Roige S, Waldman ID, Palmer AA, et al. A multivariate approach to understanding the genetic overlap between externalizing phenotypes and substance use disorders. Addict Biol. 2023;28:e13319.
Kayir H, Ruffolo J, McCunn P, Khokhar JY. The relationship between cannabis, cognition, and schizophrenia: it’s complicated. Curr Top Behav Neurosci. 2023;63:437–61.
Farrelly KN, Wardell JD, Marsden E, Scarfe ML, Najdzionek P, Turna J, et al. The impact of recreational cannabis legalization on cannabis use and associated outcomes: a systematic review. Subst Abuse. 2023;17:11782218231172054.
Fergusson DM, Boden JM, Horwood LJ. Psychosocial sequelae of cannabis use and implications for policy: findings from the Christchurch Health and Development Study. Soc Psychiatry Psychiatr Epidemiol. 2015;50:1317–26.
van der Pol P, Liebregts N, de Graaf R, Korf DJ, van den Brink W, van Laar M. Predicting the transition from frequent cannabis use to cannabis dependence: a three-year prospective study. Drug Alcohol Depend. 2013;133:352–9.
Walden N, Earleywine M. How high: quantity as a predictor of cannabis-related problems. Harm Reduct J. 2008;5:20.
Zeisser C, Thompson K, Stockwell T, Duff C, Chow C, Vallance K, et al. A ‘standard joint’? The role of quantity in predicting cannabis-related problems. Addiction Research & Theory. 2012;20:82–92.
Substance Abuse and Mental Health Services Administration. Key substance use and mental health indicators in the United States: Results from the 2022 National Survey on Drug Use and Health. Rockville, MD: Center for Behavioral Health Statistics and Quality, Substance Abuse and Mental Health Services Administration; 2022.
Althubaiti A. Information bias in health research: definition, pitfalls, and adjustment methods. J Multidiscip Healthc. 2016;9:211–7.
Jeffers AM, Glantz S, Byers A, Keyhani S. Sociodemographic characteristics associated with and prevalence and frequency of cannabis use among adults in the US. JAMA Netw Open. 2021;4:e2136571.
Karriker-Jaffe KJ. Neighborhood socioeconomic status and substance use by U.S. adults. Drug Alcohol Depend. 2013;133:212–21.
Atkinson EG, Bianchi SB, Ye GY, Martinez-Magana JJ, Tietz GE, Montalvo-Ortiz JL, et al. Cross-ancestry genomic research: time to close the gap. Neuropsychopharmacology. 2022;47:1737–8.
Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–91.
Acknowledgements
HHAT is funded by a Canadian Institute of Health Research (CIHR) Postdoctoral Fellowship (#491556). JYK is funded by a CIHR Canada Research Chair in Translational Neuropsychopharmacology. PF, the 23andMe Research Team, and SLE are employed by 23andMe, Inc. AAP is supported by NIDA (P50DA037844 and P30DA060810). SSR is funded through the National Institute on Drug Abuse (NIDA DP1DA054394) and the Tobacco-Related Disease Research Program (T32IR5226). We would like to thank the research participants and employees of 23andMe for making this work possible. Participants provided informed consent and volunteered to participate in the research online, under a protocol approved by the external AAHRPP-accredited Institutional Review Board (IRB), Ethical & Independent (E&I) Review Services. As of 2022, E&I Review Services is part of Salus IRB (https://www.versiticlinicaltrials.org/salusirb). The following members of the 23andMe Research Team contributed to this study: Stella Aslibekyan, Adam Auton, Elizabeth Babalola, Robert K. Bell, Jessica Bielenberg, Katarzyna Bryc, Emily Bullis, Daniella Coker, Gabriel Cuellar Partida, Devika Dhamija, Sayantan Das, Teresa Filshtein, Kipper Fletez-Brant, Will Freyman, Karl Heilbron, Pooja M. Gandhi, Karl Heilbron, Barry Hicks, David A. Hinds, Ethan M. Jewett, Yunxuan Jiang, Katelyn Kukar, Keng-Han Lin, Maya Lowe, Jey C. McCreight, Matthew H. McIntyre, Steven J. Micheletti, Meghan E. Moreno, Joanna L. Mountain, Priyanka Nandakumar, Elizabeth S. Noblin, Jared O’Connell, Aaron A. Petrakovitz, G. David Poznik, Morgan Schumacher, Anjali J. Shastri, Janie F. Shelton, Jingchunzi Shi, Suyash Shringarpure, Vinh Tran, Joyce Y. Tung, Xin Wang, Wei Wang, Catherine H. Weldon, Peter Wilton, Alejandro Hernandez, Corinna Wong, Christophe Toukam Tchakouté. We would also like to thank The Externalizing Consortium for sharing the GWAS summary statistics of externalizing. The Externalizing Consortium: Principal Investigators: Danielle M. Dick, Philipp Koellinger, K. Paige Harden, Abraham A. Palmer. Lead Analysts: Richard Karlsson Linnér, Travis T. Mallard, Peter B. Barr, Sandra Sanchez-Roige. Significant Contributors: Irwin D. Waldman. The Externalizing Consortium has been supported by the National Institute on Alcohol Abuse and Alcoholism (R01AA015416 -administrative supplement), and the National Institute on Drug Abuse (R01DA050721). Additional funding for investigator effort has been provided by K02AA018755, U10AA008401, P50AA022537, as well as a European Research Council Consolidator Grant (647648 EdGe to Koellinger). The content is solely the responsibility of the authors and does not necessarily represent the official views of the above funding bodies. The Externalizing Consortium would like to thank the following groups for making the research possible: 23andMe, Add Health, Vanderbilt University Medical Center’s BioVU, Collaborative Study on the Genetics of Alcoholism (COGA), the Psychiatric Genomics Consortium’s Substance Use Disorders working group, UK10K Consortium, UK Biobank, and Philadelphia Neurodevelopmental Cohort. We gratefully acknowledge All of Us participants for their contributions, without whom this research would not have been possible. We also thank the National Institutes of Health’s All of Us Research Program for making available the participant data examined in this study. The All of Us Research Program is supported by the National Institutes of Health, Office of the Director: Regional Medical Centers: 1 OT2 OD026549; 1 OT2 OD026554; 1 OT2 OD026557; 1 OT2 OD026556; 1 OT2 OD026550; 1 OT2 OD 026552; 1 OT2 OD026553; 1 OT2 OD026548; 1 OT2 OD026551; 1 OT2 OD026555; IAA #: AOD 16037; Federally Qualified Health Centers: HHSN 263201600085U; Data and Research Center: 5 U2C OD023196; Biobank: 1 U24 OD023121; The Participant Center: U24 OD023176; Participant Technology Systems Center: 1 U24 OD023163; Communications and Engagement: 3 OT2 OD023205; 3 OT2 OD023206; and Community Partners: 1 OT2 OD025277; 3 OT2 OD025315; 1 OT2 OD025337; 1 OT2 OD025276.
Author information
Authors and Affiliations
Consortia
Contributions
SSR and AAP conceived the idea. PF and SLE contributed formal analyses and curation of 23andMe data. HHAT contributed to formal analyses, investigation, and data visualization. contributed to formal data analysis and data visualization. JJM, MVJ, RBC, and SP contributed to formal analyses. HHAT and SSR wrote the manuscript. HHAT, PF, JJM, MVJ, RBC, SP, SLE, JYK, LKD, ECJ, AAP and SSR reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
PF, the 23andMe Research Team, and SLE were employed by 23andMe, Inc. PF and SLE hold stock or stock options in 23andMe, Inc. The remaining authors have nothing to disclose.
Ethics statement
All methods were performed in accordance with the relevant guidelines and regulations. Participants provided informed consent and volunteered to participate in research online under a protocol approved by the external AAHRPP-accredited Institutional Review Board (IRB), Ethical & Independent (E&I) Review Services.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Thorpe, H.H.A., Fontanillas, P., Meredith, J.J. et al. Genome-wide association studies of lifetime and frequency of cannabis use in 131,895 individuals. Mol Psychiatry 31, 1061–1073 (2026). https://doi.org/10.1038/s41380-025-03219-2
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41380-025-03219-2
This article is cited by
-
Getting into the weed—the genetics of cannabis use in modern America
Neuropsychopharmacology (2026)