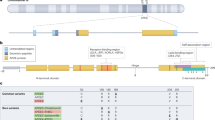
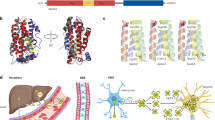
Abstract
Alzheimer’ s disease (AD) is a progressive neurodegenerative disorder characterized by a spectrum of cognitive impairments, ranging from mild memory loss to severe cognitive decline and, ultimately, death. The global incidence of AD is projected to increase significantly, with late-onset AD being predominantly sporadic in nature. Over the past three decades, the Apolipoprotein E (APOE) gene has been recognized as the most important single genetic determinant of sporadic AD risk. The APOE4 allele is a major risk factor for AD and is known to exacerbate the pathological process for AD. Identifying protective variants that may reduce the risk or delay the onset of AD is of great significance for the development of effective treatments. This review comprehensively examines the protective effects of APOE and its related protective mutations. It also explores the impact of these unique protective variants at the cellular level during the pathological progression of AD. Furthermore, the review compiles new insights for AD treatment offered by these protective mutations, exploring the potential applications of APOE and its related protective variants in advanced therapeutic strategies, including gene editing, RNA editing, and stem cell therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data analyzed during this study are available from the authors on request.
References
Andrews SJ, Fulton-Howard B, Goate A. Protective variants in Alzheimer’s disease. Curr Genet Med Rep. 2019;7:1–12.
Hurd MD, Martorell P, Delavande A, Mullen KJ, Langa KM. Monetary costs of dementia in the United States. N Engl J Med. 2013;368:1326–34.
Scheltens P, De Strooper B, Kivipelto M, Holstege H, Chételat G, Teunissen CE, et al. Alzheimer’s disease. Lancet. 2021;397:1577–90.
Vélez JI, Lopera F, Sepulveda-Falla D, Patel HR, Johar AS, Chuah A, et al. APOE*E2 allele delays age of onset in PSEN1 E280A Alzheimer’s disease. Mol Psychiatry. 2016;21:916–24.
Ayers KL, Mirshahi UL, Wardeh AH, Murray MF, Hao K, Glicksberg BS, et al. A loss of function variant in CASP7 protects against Alzheimer’s disease in homozygous APOE ε4 allele carriers. BMC Genomics. 2016;17:445.
Nelson MR, Liu P, Agrawal A, Yip O, Blumenfeld J, Traglia M, et al. The APOE-R136S mutation protects against APOE4-driven Tau pathology, neurodegeneration and neuroinflammation. Nat Neurosci. 2023;26:2104–21.
Stone DJ, Rozovsky I, Morgan TE, Anderson CP, Hajian H, Finch CE. Astrocytes and microglia respond to estrogen with increased apoE mRNA in vivo and in vitro. Exp Neurol. 1997;143:313–8.
Xu Q, Bernardo A, Walker D, Kanegawa T, Mahley RW, Huang Y. Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J Neurosci. 2006;26:4985–94.
Poirier J. Apolipoprotein E in the brain and its role in Alzheimer’s disease. J Psychiatry Neurosci. 1996;21:128–34.
Uchihara T, Duyckaerts C, He Y, Kobayashi K, Seilhean D, Amouyel P, et al. ApoE immunoreactivity and microglial cells in Alzheimer’s disease brain. Neurosci Lett. 1995;195:5–8.
Zalocusky KA, Najm R, Taubes AL, Hao Y, Yoon SY, Koutsodendris N, et al. Neuronal ApoE upregulates MHC-I expression to drive selective neurodegeneration in Alzheimer’s disease. Nat Neurosci. 2021;24:786–98.
Nilsson L, Rogers J, Potter H. The essential role of inflammation and induced gene expression in the pathogenic pathway of Alzheimer’s disease. Front Biosci. 1998;3:436–46.
Potter H, Wefes IM, Nilsson LN. The inflammation-induced pathological chaperones ACT and apo-E are necessary catalysts of Alzheimer amyloid formation. Neurobiol Aging. 2001;22:923–30.
Rozemuller AJ, van Gool WA, Eikelenboom P. The neuroinflammatory response in plaques and amyloid angiopathy in Alzheimer’s disease: therapeutic implications. Curr Drug Targets CNS Neurol Disord. 2005;4:223–33.
Potter H, Wisniewski T. Apolipoprotein e: essential catalyst of the Alzheimer amyloid cascade. Int J Alzheimers Dis. 2012;2012:489428.
Nilsson LN, Arendash GW, Leighty RE, Costa DA, Low MA, Garcia MF, et al. Cognitive impairment in PDAPP mice depends on ApoE and ACT-catalyzed amyloid formation. Neurobiol Aging. 2004;25:1153–67.
Ma J, Brewer HB Jr., Potter H. Alzheimer A beta neurotoxicity: promotion by antichymotrypsin, ApoE4; inhibition by A beta-related peptides. Neurobiol Aging. 1996;17:773–80.
Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:1977–81.
Ma J, Yee A, Brewer HB Jr., Das S, Potter H. Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature. 1994;372:92–94.
Sanan DA, Weisgraber KH, Russell SJ, Mahley RW, Huang D, Saunders A, et al. Apolipoprotein E associates with beta amyloid peptide of Alzheimer’s disease to form novel monofibrils. Isoform apoE4 associates more efficiently than apoE3. J Clin Invest. 1994;94:860–9.
Wisniewski T, Castaño EM, Golabek A, Vogel T, Frangione B:. Acceleration of Alzheimer’s fibril formation by apolipoprotein E in vitro. Am J Pathol. 1994;145:1030–5.
Deane R, Sagare A, Hamm K, Parisi M, Lane S, Finn MB, et al. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J Clin Invest. 2008;118:4002–13.
LaDu MJ, Falduto MT, Manelli AM, Reardon CA, Getz GS, Frail DE. Isoform-specific binding of apolipoprotein E to beta-amyloid. J Biol Chem. 1994;269:23403–6.
Deane R, Wu Z, Sagare A, Davis J, Du Yan S, Hamm K, et al. LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron. 2004;43:333–44.
Nilsson LN, Gografe S, Costa DA, Hughes T, Dressler D, Potter H. Use of fused circulations to investigate the role of apolipoprotein E as amyloid catalyst and peripheral sink in alzheimer’s disease. Technol Innov. 2012;14:199–208.
Arboleda-Velasquez JF, Lopera F, O’Hare M, Delgado-Tirado S, Marino C, Chmielewska N, et al. Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: a case report. Nat Med. 2019;25:1680–3.
Reitz C. Genetic diagnosis and prognosis of Alzheimer’s disease: challenges and opportunities. Expert Rev Mol Diagn. 2015;15:339–48.
Zalocusky KA, Nelson MR, Huang Y. An Alzheimer’s-disease-protective APOE mutation. Nat Med. 2019;25:1648–9.
Mhatre SD, Tsai CA, Rubin AJ, James ML, Andreasson KI. Microglial malfunction: the third rail in the development of Alzheimer’s disease. Trends Neurosci. 2015;38:621–36.
Masters CL, Bateman R, Blennow K, Rowe CC, Sperling RA, Cummings JL. Alzheimer’s disease. Nat Rev Dis Primers. 2015;1:15056.
Rauch JN, Luna G, Guzman E, Audouard M, Challis C, Sibih YE, et al. LRP1 is a master regulator of tau uptake and spread. Nature. 2020;580:381–5.
Ji ZS, Pitas RE, Mahley RW. Differential cellular accumulation/retention of apolipoprotein E mediated by cell surface heparan sulfate proteoglycans. Apolipoproteins E3 and E2 greater than e4. J Biol Chem. 1998;273:13452–60.
Le Guen Y, Belloy ME, Grenier-Boley B, de Rojas I, Castillo-Morales A, Jansen I, et al. Association of rare APOE missense variants V236E and R251G with risk of alzheimer disease. JAMA Neurol. 2022;79:652–63.
Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–3.
Tang MX, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, et al. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998;279:751–5.
Huang Y, Mahley RW. Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol Dis. 2014;72:3–12.
Zhao J, Fu Y, Yamazaki Y, Ren Y, Davis MD, Liu CC, et al. APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer’s disease patient iPSC-derived cerebral organoids. Nat Commun. 2020;11:5540.
Bhattarai P, Gunasekaran TI, Belloy ME, Reyes-Dumeyer D, Jülich D, Tayran H, et al. Rare genetic variation in fibronectin 1 (FN1) protects against APOEε4 in Alzheimer’s disease. Acta Neuropathol. 2024;147:70.
Chemparathy A, Le Guen Y, Chen S, Lee EG, Leong L, Gorzynski JE, et al. APOE loss-of-function variants: Compatible with longevity and associated with resistance to Alzheimer’s disease pathology. Neuron. 2024;112:1110–e1115.
Lin YT, Seo J, Gao F, Feldman HM, Wen HL, Penney J, et al. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron. 2018;98:1141–e1147.
Belloy ME, Napolioni V, Han SS, Le Guen Y, Greicius MD. Association of klotho-VS heterozygosity with risk of Alzheimer disease in individuals who carry APOE4. JAMA Neurol. 2020;77:849–62.
Quiroz YT, Aguillon D, Aguirre-Acevedo DC, Vasquez D, Zuluaga Y, Baena AY, et al. APOE3 christchurch heterozygosity and autosomal dominant Alzheimer’s disease. N Engl J Med. 2024;390:2156–64.
Mahley RW, Huang Y, Rall SC Jr. Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia). Questions, quandaries, and paradoxes. J Lipid Res. 1999;40:1933–49.
Naguib S, Lopez-Lee C, Torres ER, Lee SI, Zhu J, Zhu D, et al. The R136S mutation in the APOE3 gene confers resilience against tau pathology via inhibition of the cGAS-STING-IFN pathway. Immunity 2025;58:1931–47.e1939.
Wardell MR, Brennan SO, Janus ED, Fraser R, Carrell RW. Apolipoprotein E2-Christchurch (136 Arg-Ser). New variant of human apolipoprotein E in a patient with type III hyperlipoproteinemia. J Clin Invest. 1987;80:483–90.
Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–59.
Günaydin C, Sondhi D, Kaminsky SM, Lephart HC, Leopold PL, Hackett NR, et al. AAVrh.10 delivery of novel APOE2-christchurch variant suppresses amyloid and Tau pathology in Alzheimer’s disease mice. Mol Ther. 2024;32:4303–18.
Rauch JN, Chen JJ, Sorum AW, Miller GM, Sharf T, See SK, et al. Tau internalization is regulated by 6-O sulfation on heparan sulfate proteoglycans (HSPGs). Sci Rep. 2018;8:6382.
Chen Y, Song S, Parhizkar S, Lord J, Zhu Y, Strickland MR, et al. APOE3ch alters microglial response and suppresses Aβ-induced tau seeding and spread. Cell. 2024;187:428–e420.
Tian X, Rivera EK, Hwang I, Nikitina AA, Smith JA, Cho YJ, et al. Protective mechanisms against Alzheimer’s disease in APOE3-Christchurch homozygous astrocytes. Alzheimers Dement. 2025;21:e70589.
Simonovitch S, Schmukler E, Bespalko A, Iram T, Frenkel D, Holtzman DM, et al. Impaired Autophagy in APOE4 Astrocytes. J Alzheimers Dis. 2016;51:915–27.
Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer’s disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol. 2000;157:277–86.
Ji ZS, Miranda RD, Newhouse YM, Weisgraber KH, Huang Y, Mahley RW. Apolipoprotein E4 potentiates amyloid beta peptide-induced lysosomal leakage and apoptosis in neuronal cells. J Biol Chem. 2002;277:21821–8.
Shi Y, Andhey PS, Ising C, Wang K, Snipes LL, Boyer K, et al. Overexpressing low-density lipoprotein receptor reduces tau-associated neurodegeneration in relation to apoE-linked mechanisms. Neuron. 2021;109:2413–e2417.
Tran KM, Kwang NE, Butler CA, Gomez-Arboledas A, Kawauchi S, Mar C, et al. APOE Christchurch enhances a disease-associated microglial response to plaque but suppresses response to tau pathology. Mol Neurodegener. 2025;20:9.
Futamura M, Dhanasekaran P, Handa T, Phillips MC, Lund-Katz S, Saito H. Two-step mechanism of binding of apolipoprotein E to heparin: implications for the kinetics of apolipoprotein E-heparan sulfate proteoglycan complex formation on cell surfaces. J Biol Chem. 2005;280:5414–22.
Moon HJ, Luo Y, Chugh D, Zhao L. Human apolipoprotein E glycosylation and sialylation: from structure to function. Front Mol Neurosci. 2024;17:1399965.
Chen J, Li Q, Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc Natl Acad Sci USA. 2011;108:14813–8.
Lalazar A, Weisgraber KH, Rall SC Jr., Giladi H, Innerarity TL, Levanon AZ, et al. Site-specific mutagenesis of human apolipoprotein E. Receptor binding activity of variants with single amino acid substitutions. J Biol Chem. 1988;263:3542–5.
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–w303.
Perez-Corredor P, Vanderleest TE, Vacano GN, Sanchez JS, Villalba-Moreno ND, Marino C, et al. APOE3 Christchurch modulates β-catenin/Wnt signaling in iPS cell-derived cerebral organoids from Alzheimer’s cases. Front Mol Neurosci. 2024;17:1373568.
Littau JL, Velilla L, Hase Y, Villalba-Moreno ND, Hagel C, Drexler D, et al. Evidence of beta amyloid independent small vessel disease in familial Alzheimer’s disease. Brain Pathol. 2022;32:e13097.
Kisler K, Nelson AR, Montagne A, Zlokovic BV. Cerebral blood flow regulation and neurovascular dysfunction in Alzheimer disease. Nat Rev Neurosci. 2017;18:419–34.
Liedtke W, Edelmann W, Bieri PL, Chiu FC, Cowan NJ, Kucherlapati R, et al. GFAP is necessary for the integrity of CNS white matter architecture and long-term maintenance of myelination. Neuron. 1996;17:607–15.
Zhao Z, Nelson AR, Betsholtz C, Zlokovic BV. Establishment and dysfunction of the blood-brain barrier. Cell. 2015;163:1064–78.
Obermeier B, Daneman R, Ransohoff RM. Development, maintenance and disruption of the blood-brain barrier. Nat Med. 2013;19:1584–96.
Zlokovic BV. Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nat Rev Neurosci. 2011;12:723–38.
Sweeney MD, Sagare AP, Zlokovic BV. Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat Rev Neurol. 2018;14:133–50.
Dave JM, Bayless KJ. Vimentin as an integral regulator of cell adhesion and endothelial sprouting. Microcirculation. 2014;21:333–44.
Sullivan SM, Lee A, Björkman ST, Miller SM, Sullivan RK, Poronnik P, et al. Cytoskeletal anchoring of GLAST determines susceptibility to brain damage: an identified role for GFAP. J Biol Chem. 2007;282:29414–23.
Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:1–105.
Roberts RC, Roche JK, McCullumsmith RE. Localization of excitatory amino acid transporters EAAT1 and EAAT2 in human postmortem cortex: a light and electron microscopic study. Neuroscience. 2014;277:522–40.
Armbruster M, Hanson E, Dulla CG. Glutamate clearance is locally modulated by presynaptic neuronal activity in the cerebral cortex. J Neurosci. 2016;36:10404–15.
Henao-Restrepo J, López-Murillo C, Valderrama-Carmona P, Orozco-Santa N, Gomez J, Gutiérrez-Vargas J, et al. Gliovascular alterations in sporadic and familial Alzheimer’s disease: APOE3 Christchurch homozygote glioprotection. Brain Pathol. 2023;33:e13119.
Hashimoto T, Serrano-Pozo A, Hori Y, Adams KW, Takeda S, Banerji AO, et al. Apolipoprotein E, especially apolipoprotein E4, increases the oligomerization of amyloid β peptide. J Neurosci. 2012;32:15181–92.
Fersht A: Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding: Macmillan; 1999.
Aleshkov S, Abraham CR, Zannis VI. Interaction of nascent ApoE2, ApoE3, and ApoE4 isoforms expressed in mammalian cells with amyloid peptide beta (1-40). Relevance to Alzheimer’s disease. Biochemistry. 1997;36:10571–80.
Tokuda T, Calero M, Matsubara E, Vidal R, Kumar A, Permanne B, et al. Lipidation of apolipoprotein E influences its isoform-specific interaction with Alzheimer’s amyloid beta peptides. Biochem J. 2000;348:359–65.
Lin PB, Holtzman DM. Current insights into apolipoprotein E and the immune response in Alzheimer’s disease. Immunol Rev. 2024;327:43–52.
Medway CW, Abdul-Hay S, Mims T, Ma L, Bisceglio G, Zou F, et al. ApoE variant p.V236E is associated with markedly reduced risk of Alzheimer’s disease. Mol Neurodegener. 2014;9:11.
Liu CC, Murray ME, Li X, Zhao N, Wang N, Heckman MG, et al. APOE3-Jacksonville (V236E) variant reduces self-aggregation and risk of dementia. Sci Transl Med. 2021;13:eabc9375.
Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–54.
Lund-Katz S, Phillips MC. High density lipoprotein structure-function and role in reverse cholesterol transport. Subcell Biochem. 2010;51:183–227.
Choy N, Raussens V, Narayanaswami V. Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain. J Mol Biol. 2003;334:527–39.
Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10:333–44.
Bales KR, Verina T, Dodel RC, Du Y, Altstiel L, Bender M, et al. Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat Genet. 1997;17:263–4.
Ulrich JD, Ulland TK, Mahan TE, Nyström S, Nilsson KP, Song WM, et al. ApoE facilitates the microglial response to amyloid plaque pathology. J Exp Med. 2018;215:1047–58.
Bu G. APOE targeting strategy in Alzheimer’s disease: lessons learned from protective variants. Mol Neurodegener. 2022;17:51.
Huang Y, Weisgraber KH, Mucke L, Mahley RW. Apolipoprotein E: diversity of cellular origins, structural and biophysical properties, and effects in Alzheimer’s disease. J Mol Neurosci. 2004;23:189–204.
Chouraki V, Seshadri S. Genetics of Alzheimer’s disease. Adv Genet. 2014;87:245–94.
Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, et al. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry. 2011;16:903–7.
Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. JAMA. 1997;278:1349–56. APOE and Alzheimer Disease Meta Analysis Consortium
Narasimhappagari J, Liu L, Balasubramaniam M, Ayyadevara S, Griffin WST. When two worlds collide: the contribution and association between genetics (APOEε4) and neuroinflammation (IL-1β) in Alzheimer’s Neuropathogenesis. Cells. 2025;14:1216.
Balasubramaniam M, Narasimhappagari J, Liu L, Ganne A, Ayyadevara S, Atluri R, et al. Rescue of ApoE4-related lysosomal autophagic failure in Alzheimer’s disease by targeted small molecules. Commun Biol. 2024;7:60.
Li C, Götz J. Tau-based therapies in neurodegeneration: opportunities and challenges. Nat Rev Drug Discov. 2017;16:863–83.
Agosta F, Vossel KA, Miller BL, Migliaccio R, Bonasera SJ, Filippi M, et al. Apolipoprotein E epsilon4 is associated with disease-specific effects on brain atrophy in Alzheimer’s disease and frontotemporal dementia. Proc Natl Acad Sci USA. 2009;106:2018–22.
Shi Y, Yamada K, Liddelow SA, Smith ST, Zhao L, Luo W, et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. 2017;549:523–7.
Therriault J, Benedet AL, Pascoal TA, Mathotaarachchi S, Chamoun M, Savard M, et al. Association of apolipoprotein E ε4 with medial temporal tau independent of amyloid-β. JAMA Neurol. 2020;77:470–9.
Farhy-Tselnicker I, van Casteren ACM, Lee A, Chang VT, Aricescu AR, Allen NJ. Astrocyte-secreted glypican 4 regulates release of neuronal pentraxin 1 from axons to induce functional synapse formation. Neuron. 2017;96:428–e413.
Allen NJ, Bennett ML, Foo LC, Wang GX, Chakraborty C, Smith SJ, et al. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486:410–4.
Karran E, Mercken M, De Strooper B. The amyloid cascade hypothesis for Alzheimer’s disease: an appraisal for the development of therapeutics. Nat Rev Drug Discov. 2011;10:698–712.
Shvetcov A, Johnson ECB, Winchester LM, Walker KA, Wilkins HM, Thompson TG, et al. APOE ε4 carriers share immune-related proteomic changes across neurodegenerative diseases. Nat Med. 2025;31:2590–601.
Shang Z, Lv H, Zhang M, Duan L, Wang S, Li J, et al. Genome-wide haplotype association study identify TNFRSF1A, CASP7, LRP1B, CDH1 and TG genes associated with Alzheimer’s disease in Caribbean Hispanic individuals. Oncotarget. 2015;6:42504–14.
Gurland BJ, Wilder DE, Lantigua R, Stern Y, Chen J, Killeffer EH, et al. Rates of dementia in three ethnoracial groups. Int J Geriatr Psychiatry. 1999;14:481–93.
Burguillos MA, Deierborg T, Kavanagh E, Persson A, Hajji N, Garcia-Quintanilla A, et al. Caspase signalling controls microglia activation and neurotoxicity. Nature. 2011;472:319–24.
Hansen DV, Hanson JE, Sheng M. Microglia in Alzheimer’s disease. J Cell Biol. 2018;217:459–72.
Szekely CA, Breitner JC, Fitzpatrick AL, Rea TD, Psaty BM, Kuller LH, et al. NSAID use and dementia risk in the cardiovascular health study: role of APOE and NSAID type. Neurology. 2008;70:17–24.
Schumacher S, Dedden D, Nunez RV, Matoba K, Takagi J, Biertümpfel C, et al. Structural insights into integrin α(5)β(1) opening by fibronectin ligand. Sci Adv. 2021;7:eabe9716.
Michalski D, Spielvogel E, Puchta J, Reimann W, Barthel H, Nitzsche B, et al. Increased immunosignals of collagen IV and fibronectin indicate ischemic consequences for the neurovascular matrix adhesion zone in various animal models and human stroke tissue. Front Physiol. 2020;11:575598.
Patel T, Carnwath TP, Wang X, Allen M, Lincoln SJ, Lewis-Tuffin LJ, et al. Transcriptional landscape of human microglia implicates age, sex, and APOE-related immunometabolic pathway perturbations. Aging Cell. 2022;21:e13606.
Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004;5:146–56.
Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, et al. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32:6391–410.
Bellaver B, Povala G, Ferreira PCL, Ferrari-Souza JP, Leffa DT, Lussier FZ, et al. Astrocyte reactivity influences amyloid-β effects on tau pathology in preclinical Alzheimer’s disease. Nat Med. 2023;29:1775–81.
Lee AJ, Raghavan NS, Bhattarai P, Siddiqui T, Sariya S, Reyes-Dumeyer D, et al. FMNL2 regulates gliovascular interactions and is associated with vascular risk factors and cerebrovascular pathology in Alzheimer’s disease. Acta Neuropathol. 2022;144:59–79.
Heneka MT, Carson MJ, El Khoury J, Landreth GE, Brosseron F, Feinstein DL, et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015;14:388–405.
Paresce DM, Ghosh RN, Maxfield FR. Microglial cells internalize aggregates of the Alzheimer’s disease amyloid beta-protein via a scavenger receptor. Neuron. 1996;17:553–65.
Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–33.
Arking DE, Krebsova A, Macek M Sr., Macek M Jr., Arking A, Mian IS, et al. Association of human aging with a functional variant of klotho. Proc Natl Acad Sci USA. 2002;99:856–61.
Ullah M, Sun Z. Klotho deficiency accelerates stem cells aging by impairing telomerase activity. J Gerontol A Biol Sci Med Sci. 2019;74:1396–407.
Fagan AM, Mintun MA, Mach RH, Lee SY, Dence CS, Shah AR, et al. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol. 2006;59:512–9.
Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, et al. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol. 2010;67:122–31.
Jack CR Jr., Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–16.
Burnham SC, Bourgeat P, Doré V, Savage G, Brown B, Laws S, et al. Clinical and cognitive trajectories in cognitively healthy elderly individuals with suspected non-Alzheimer’s disease pathophysiology (SNAP) or Alzheimer’s disease pathology: a longitudinal study. Lancet Neurol. 2016;15:1044–53.
Vos SJ, Xiong C, Visser PJ, Jasielec MS, Hassenstab J, Grant EA, et al. Preclinical Alzheimer’s disease and its outcome: a longitudinal cohort study. Lancet Neurol. 2013;12:957–65.
Ossenkoppele R, Jansen WJ, Rabinovici GD, Knol DL, van der Flier WM, van Berckel BN, et al. Prevalence of amyloid PET positivity in dementia syndromes: a meta-analysis. JAMA. 2015;313:1939–49.
Lim YY, Kalinowski P, Pietrzak RH, Laws SM, Burnham SC, Ames D, et al. Association of β-amyloid and apolipoprotein E ε4 with memory decline in preclinical alzheimer disease. JAMA Neurol. 2018;75:488–94.
Li H, Wang B, Wang Z, Guo Q, Tabuchi K, Hammer RE, et al. Soluble amyloid precursor protein (APP) regulates transthyretin and Klotho gene expression without rescuing the essential function of APP. Proc Natl Acad Sci USA. 2010;107:17362–7.
O’Brien RJ, Wong PC. Amyloid precursor protein processing and Alzheimer’s disease. Annu Rev Neurosci. 2011;34:185–204.
Chen CD, Podvin S, Gillespie E, Leeman SE, Abraham CR. Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proc Natl Acad Sci USA. 2007;104:19796–801.
Bloch L, Sineshchekova O, Reichenbach D, Reiss K, Saftig P, Kuro-o M, et al. Klotho is a substrate for alpha-, beta- and gamma-secretase. FEBS Lett. 2009;583:3221–4.
Inestrosa NC, Tapia-Rojas C, Lindsay CB, Zolezzi JM. Wnt signaling pathway dysregulation in the aging brain: lessons from the octodon degus. Front Cell Dev Biol. 2020;8:734.
Sadowski M, Pankiewicz J, Scholtzova H, Ripellino JA, Li Y, Schmidt SD, et al. A synthetic peptide blocking the apolipoprotein E/beta-amyloid binding mitigates beta-amyloid toxicity and fibril formation in vitro and reduces beta-amyloid plaques in transgenic mice. Am J Pathol. 2004;165:937–48.
Johnson NR, Wang AC, Coughlan C, Sillau S, Lucero E, Viltz L, et al. Imipramine and olanzapine block apoE4-catalyzed polymerization of Aβ and show evidence of improving Alzheimer’s disease cognition. Alzheimers Res Ther. 2022;14:88.
Wang C, Najm R, Xu Q, Jeong DE, Walker D, Balestra ME, et al. Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat Med. 2018;24:647–57.
Rottner AK, Lundin A, Li S, Firth M, Maresca M, Sienski G. Optimized prime editing of the Alzheimer’s disease-associated APOE4 mutation. Stem Cell Reports. 2025;20:102372.
Kantor A, McClements ME, MacLaren RE. CRISPR-Cas9 DNA Base-Editing and Prime-Editing. Int J Mol Sci. 2020;21:6240.
MacKenzie SH, Schipper JL, Clark AC. The potential for caspases in drug discovery. Curr Opin Drug Discov Devel. 2010;13:568–76.
Golden LR, Siano DS, Stephens IO, MacLean SM, Saito K, Nolt GL, et al. APOE4 to APOE2 allelic switching in mice improves Alzheimer’s disease-related metabolic signatures, neuropathology and cognition. Nat Neurosci. 2025.
Xu Z, Wang Q, Zhong H, Jiang Y, Shi X, Yuan B, et al. Carrier strategies boost the application of CRISPR/Cas system in gene therapy. Exploration (Beijing). 2022;2:20210081.
Tang W, Liu J, Ding B. Nucleic acid nanostructure for delivery of CRISPR/Cas9‐based gene editing system. Interdisciplinary Medicine. 2023;1:e20220014.
Blumenfeld J, Yip O, Kim MJ, Huang Y. Cell type-specific roles of APOE4 in Alzheimer disease. Nat Rev Neurosci. 2024;25:91–110.
Lenharo M. Move over, CRISPR: RNA-editing therapies pick up steam. Nature. 2024;626:933–4.
Balez R, Steiner N, Engel M, Muñoz SS, Lum JS, Wu Y, et al. Neuroprotective effects of apigenin against inflammation, neuronal excitability and apoptosis in an induced pluripotent stem cell model of Alzheimer’s disease. Sci Rep. 2016;6:31450.
Hossini AM, Megges M, Prigione A, Lichtner B, Toliat MR, Wruck W, et al. Induced pluripotent stem cell-derived neuronal cells from a sporadic Alzheimer’s disease donor as a model for investigating AD-associated gene regulatory networks. BMC Genomics. 2015;16:84.
Muratore CR, Rice HC, Srikanth P, Callahan DG, Shin T, Benjamin LN, et al. The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet. 2014;23:3523–36.
Phillips MI, Tang YL. Genetic modification of stem cells for transplantation. Adv Drug Deliv Rev. 2008;60:160–72.
Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–86.
Yan Y, Tao H, He J, Huang SY. The HDOCK server for integrated protein-protein docking. Nat Protoc. 2020;15:1829–52.
UniProt Consortium. UniProt: the universal protein knowledgebase in 2025. Nucleic Acids Res. 2025;53:D609–d617.
Teufel F, Almagro Armenteros JJ, Johansen AR, Gíslason MH, Pihl SI, Tsirigos KD, et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol. 2022;40:1023–5.
B. Rupp BWS, M. Forstner: Apolipoprotein E3 (APOE3) truncation mutant 165. In.; 1999.
Verderame JR, Kantardjieff K., Segelke B, Weisgraber K, Rupp B Apolipoprotein E4, 22k domain. In. 2002.
Jeon H, Meng W, Takagi J, Eck MJ, Springer TA, Blacklow SC Crystal Structure of the LDL Receptor YWTD-EGF Domain Pair In. 2001.
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–42.
Lemere CA, Lopera F, Kosik KS, Lendon CL, Ossa J, Saido TC, et al. The E280A presenilin 1 Alzheimer mutation produces increased A beta 42 deposition and severe cerebellar pathology. Nat Med. 1996;2:1146–50.
Sepulveda-Falla D, Chavez-Gutierrez L, Portelius E, Vélez JI, Dujardin S, Barrera-Ocampo A, et al. A multifactorial model of pathology for age of onset heterogeneity in familial Alzheimer’s disease. Acta Neuropathol. 2021;141:217–33.
Sayed FA, Kodama L, Fan L, Carling GK, Udeochu JC, Le D, et al. AD-linked R47H-TREM2 mutation induces disease-enhancing microglial states via AKT hyperactivation. Sci Transl Med. 2021;13:eabe3947.
Harris FM, Brecht WJ, Xu Q, Mahley RW, Huang Y. Increased tau phosphorylation in apolipoprotein E4 transgenic mice is associated with activation of extracellular signal-regulated kinase: modulation by zinc. J Biol Chem. 2004;279:44795–801.
Acknowledgements
We acknowledge the participants and investigators for all data sources. Figures in this manuscript were created with FigDraw.com.
Funding
This work was supported by The National Natural Science Foundation of China (82305298), The Fundamental Research Funds for the Central Universities (2025-JYB-XJSJJ037), The Youth Talent Support Program of the China Association of Chinese Medicine (2024-2026) (CACM-2024-QNRC2-B36), The Central High-level Hospital of Traditional Chinese Medicine: Beijing University of Traditional Chinese Medicine Dongzhimen Hospital Talent Training Program-Youth Reserve Talent Project (DZMG-QNHB0010), Beijing University of Chinese Medicine Dongzhimen Hospital Clinical research and achievement transformation ability improvement project-Youth special project (DZMG-QNZX-24003), China Postdoctoral Science Foundation funded project (2022M720521), Young Elite Scientists Sponsorship Program of the Beijing High Innovation Plan (No.20250916, No.2025KXQT03).
Author information
Authors and Affiliations
Contributions
Writing – original draft: Yuejia Ma, Yanxi Li and Guangrun Wu. Resources: Lulu Liu, Mengjie Tian, Xinyu Han, Xinyi Chen, Xiuchen Xuan, Tianhu Zheng, Xu Gao. Writing–review & editing: Fuyuan Li. Project administration, Funding acquisition and Writing–review & editing: Qing Xia. Project administration and Writing–review & editing: Dayong Wang. All authors read and approved the final manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, Y., Li, Y., Wu, G. et al. Protective mutations associated with APOE in Alzheimer’s disease. Mol Psychiatry (2026). https://doi.org/10.1038/s41380-026-03496-5
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41380-026-03496-5