Abstract
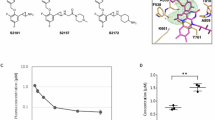
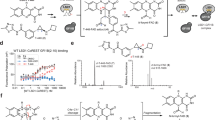
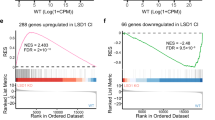
Dysregulation of histone H3 lysine 4 (H3K4) methylation has been implicated in the pathogenesis of several neurodevelopmental disorders. Targeting lysine-specific demethylase 1 (LSD1), an H3K4 demethylase, is therefore a promising approach to treat these disorders. However, LSD1 forms complexes with cofactors including growth factor independent 1B (GFI1B), a critical regulator of hematopoietic differentiation. Known tranylcypromine-based irreversible LSD1 inhibitors bind to coenzyme flavin adenine dinucleotide (FAD) and disrupt the LSD1-GFI1B complex, which is associated with hematotoxicity such as thrombocytopenia, representing a major hurdle in the development of LSD1 inhibitors as therapeutic agents. To discover LSD1 inhibitors with potent epigenetic modulation and lower risk of hematotoxicity, we screened small molecules that enhance H3K4 methylation by the inhibition of LSD1 enzyme activity in primary cultured rat neurons but have little impact on LSD1-GFI1B complex in human TF-1a erythroblasts. Here we report the discovery of a specific inhibitor of LSD1 enzyme activity, T-448 (3-((1S,2R)-2-(cyclobutylamino)cyclopropyl)-N-(5-methyl-1,3,4-thiadiazol-2-yl)benzamide fumarate). T-448 has minimal impact on the LSD1-GFI1B complex and a superior hematological safety profile in mice via the generation of a compact formyl-FAD adduct. T-448 increased brain H3K4 methylation and partially restored learning function in mice with NMDA receptor hypofunction. T-448-type LSD1 inhibitors with improved safety profiles may provide unique therapeutic approaches for central nervous system disorders associated with epigenetic dysregulation.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Cortes-Mendoza J, Diaz de Leon-Guerrero S, Pedraza-Alva G, Perez-Martinez L. Shaping synaptic plasticity: the role of activity-mediated epigenetic regulation on gene transcription. Int J Dev Neurosci. 2013;31:359–69.
Guan JS, Xie H, Ding X. The role of epigenetic regulation in learning and memory. Exp Neurol. 2015;268:30–36.
Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–99.
Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, et al. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci. 2007a;27:11254–62.
Network, Pathway Analysis Subgroup of Psychiatric Genomics C. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209.
Shulha HP, Cheung I, Whittle C, Wang J, Virgil D, Lin CL, et al. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Arch General Psychiatry. 2012;69:314–24.
Wynder C, Stalker L, Doughty ML. Role of H3K4 demethylases in complex neurodevelopmental diseases. Epigenomics. 2010;2:407–18.
Parkel S, Lopez-Atalaya JP, Barco A. Histone H3 lysine methylation in cognition and intellectual disability disorders. Learn Mem. 2013;20:570–9.
Vashishtha M, Ng CW, Yildirim F, Gipson TA, Kratter IH, Bodai L, et al. Targeting H3K4 trimethylation in Huntington disease. Proc Natl Acad Sci USA 2013;110: E3027–3036.
Schmitt ML, Ladwein KI, Carlino L, Schulz-Fincke J, Willmann D, Metzger E, et al. Heterogeneous Antibody-based Activity Assay For Lysine Specific Demethylase 1 (LSD1) on a histone peptide substrate. J Biomol Screen. 2014;19:973–8.
Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9.
Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–9.
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53.
Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol. 2006;13:626–32.
Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, et al. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat Struct Mol Biol. 2007;14:535–9.
Forneris F, Binda C, Dall’Aglio A, Fraaije MW, Battaglioli E, Mattevi A. A highly specific mechanism of histone H3-K4 recognition by histone demethylase LSD1. J Biol Chem. 2006;281:35289–95.
Wang X, Huang B, Suzuki T, Liu X, Zhan P. Medicinal chemistry insights in the discovery of novel LSD1 inhibitors. Epigenomics. 2015;7:1379–96.
Baron R, Binda C, Tortorici M, McCammon JA, Mattevi A. Molecular mimicry and ligand recognition in binding and catalysis by the histone demethylase LSD1-CoREST complex. Structure. 2011;19:212–20.
Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–72.
Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–7.
Moroy T, Vassen L, Wilkes B, Khandanpour C. From cytopenia to leukemia: the role of Gfi1 and Gfi1b in blood formation. Blood. 2015;126:2561–9.
Sprussel A, Schulte JH, Weber S, Necke M, Handschke K, Thor T, et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia. 2012;26:2039–51.
Ishikawa Y, Gamo K, Yabuki M, Takagi S, Toyoshima K, Nakayama K, et al. A novel LSD1 inhibitor T-3775440 disrupts GFI1B-containing complex leading to transdifferentiation and impaired growth of AML cells. Mol Cancer Ther. 2017;16:273–84.
Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell. 2015;28:57–69.
Rivers A, Vaitkus K, Ruiz MA, Ibanez V, Jagadeeswaran R, Kouznetsova T, et al. RN-1, a potent and selective lysine-specific demethylase 1 inhibitor, increases gamma-globin expression, F reticulocytes, and F cells in a sickle cell disease mouse model. Exp Hematol. 2015;43:546–53 e541-543.
Maiques-Diaz A, Spencer GJ, Lynch JT, Ciceri F, Williams EL, Amaral FMR, et al. Enhancer activation by pharmacologic displacement of LSD1 from GFI1 induces differentiation in acute myeloid leukemia. Cell Rep. 2018;22:3641–59.
Yamamoto R, Kawahara M, Ito S, Satoh J, Tatsumi G, Hishizawa M, et al. Selective dissociation between LSD1 and GFI1B by a LSD1 inhibitor NCD38 induces the activation of ERG super-enhancer in erythroleukemia cells. Oncotarget. 2018;9:21007–21.
Halene TB, Ehrlichman RS, Liang Y, Christian EP, Jonak GJ, Gur TL, et al. Assessment of NMDA receptor NR1 subunit hypofunction in mice as a model for schizophrenia. Genes Brain Behav. 2009;8:661–75.
Mohn AR, Gainetdinov RR, Caron MG, Koller BH. Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell. 1999;98:427–36.
Wesseling H, Guest PC, Lee CM, Wong EH, Rahmoune H, Bahn S. Integrative proteomic analysis of the NMDA NR1 knockdown mouse model reveals effects on central and peripheral pathways associated with schizophrenia and autism spectrum disorders. Mol Autism. 2014;5:38.
Kunugi A, Tajima Y, Kuno H, Sogabe S, Kimura H. HBT1, a novel AMPA receptor potentiator with lower agonistic effect, avoided bell-shaped response in in vitro BDNF production. J Pharmacol Exp Ther. 2018;364:377–89.
Tomita N, Kajii S, Douglas RC, Tomita D, Imamura S, Tsuchida K, et al. Cyclopropanamine compound. Patent WO2013022047. 2013.
Rotili D, Mai A. Targeting histone demethylases: a new avenue for the fight against cancer. Genes Cancer. 2011;2:663–79.
Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–59.
Boulle F, van den Hove DL, Jakob SB, Rutten BP, Hamon M, van Os J, et al. Epigenetic regulation of the BDNF gene: implications for psychiatric disorders. Mol Psychiatry. 2012;17:584–96.
Carter SD, Mifsud KR, Reul JM. Distinct epigenetic and gene expression changes in rat hippocampal neurons after Morris water maze training. Front Behav Neurosci. 2015;9:156.
Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–90.
Meaney MJ. Epigenetics and the biological definition of gene x environment interactions. Child Dev. 2010;81:41–79.
Tordjman S, Somogyi E, Coulon N, Kermarrec S, Cohen D, Bronsard G, et al. Gene x Environment interactions in autism spectrum disorders: role of epigenetic mechanisms. Front Psychiatry. 2014;5:53.
van Vliet J, Oates NA, Whitelaw E. Epigenetic mechanisms in the context of complex diseases. Cell Mol life Sci. 2007;64:1531–8.
Barnard RA, Pomaville MB, O’Roak BJ. Mutations and modeling of the chromatin remodeler CHD8 define an emerging autism etiology. Front Neurosci. 2015;9:477.
Katayama Y, Nishiyama M, Shoji H, Ohkawa Y, Kawamura A, Sato T, et al. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537:675–9.
Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007b;449:105–8.
Irwin RE, Pentieva K, Cassidy T, Lees-Murdock DJ, McLaughlin M, Prasad G, et al. The interplay between DNA methylation, folate and neurocognitive development. Epigenomics. 2016;8:863–79.
Papiol S, Arias B, Barrantes-Vidal N, Guitart M, Salgado P, Catalan R, et al. Analysis of polymorphisms at the tumor suppressor genep53 (TP53) in contributing to the risk for schizophrenia and its associated neurocognitive deficits. Neurosci Lett. 2004;363:78–80.
Acknowledgements
The authors thank Dr. Takayuki Niimura and Mr. Ryota Maeda for performing the pharmacological analysis, Mr. Yasushi Hattori and Dr. Shigemitsu Matsumoto for compound synthesis, Mr. Takashi Ito and Ms. Yumi Zama for protein purification, as well as Dr. Gyorgy Snell and Dr. Lane Scott for data collection and processing for crystal structure analysis. This work was funded by the Takeda Pharmaceutical Company Limited.
Author contributions
S.Matsuda and H.Kimura conceived and designed the experiments. S.Matsuda, R.B., H.O., S.Igaki, Y.K., S.Iwasaki, K.M., R.Hibino, H.Kamada, T.H., M.Iwatani, K.T., and R.Hara performed the experiments and analyzed the data. S.Morimoto, M.T., and M.Ito designed and synthesized T-448. S.Matsuda and H.Kimura wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors are present employees of the Takeda Pharmaceutical Company Limited or were employees of Takeda Pharmaceutical Company Limited while engaged in this research.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Matsuda, S., Baba, R., Oki, H. et al. T-448, a specific inhibitor of LSD1 enzyme activity, improves learning function without causing thrombocytopenia in mice. Neuropsychopharmacol. 44, 1505–1512 (2019). https://doi.org/10.1038/s41386-018-0300-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41386-018-0300-9
This article is cited by
-
Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance
Nature Communications (2025)
-
LPS-induced neuroinflammation induces changes in the transcriptional profile of members of the CoRest repressive complex in the hippocampus
Molecular Biology Reports (2024)
-
The progress of research on histone methylation in ischemic stroke pathogenesis
Journal of Physiology and Biochemistry (2022)
-
First-in-Human Randomized Trial to Assess Safety, Tolerability, Pharmacokinetics and Pharmacodynamics of the KDM1A Inhibitor Vafidemstat
CNS Drugs (2021)